Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charcot-Marie-Tooth neuropathy
- PMID: 20178975
- PMCID: PMC2857091
- DOI: 10.1074/jbc.M109.082644
Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charcot-Marie-Tooth neuropathy
Abstract
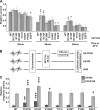
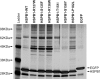
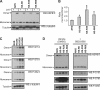
Small heat shock proteins are molecular chaperones capable of maintaining denatured proteins in a folding-competent state. We have previously shown that missense mutations in the small heat shock protein HSPB1 (HSP27) cause distal hereditary motor neuropathy and axonal Charcot-Marie-Tooth disease. Here we investigated the biochemical consequences of HSPB1 mutations that are known to cause peripheral neuropathy. In contrast to other chaperonopathies, our results revealed that particular HSPB1 mutations presented higher chaperone activity compared with wild type. Hyperactivation of HSPB1 was accompanied by a change from its wild-type dimeric state to a monomer without dissociation of the 24-meric state. Purification of protein complexes from wild-type and HSPB1 mutants showed that the hyperactive isoforms also presented enhanced binding to client proteins. Furthermore, we show that the wild-type HSPB1 protein undergoes monomerization during heat-shock activation, strongly suggesting that the monomer is the active form of the HSPB1 protein.
Figures


References
-
- Barisic N., Claeys K. G., Sirotković-Skerlev M., Löfgren A., Nelis E., De Jonghe P., Timmerman V. (2008) Ann. Hum. Genet. 72, 416–441 - PubMed
-
- Evgrafov O. V., Mersiyanova I., Irobi J., Van Den Bosch L., Dierick I., Leung C. L., Schagina O., Verpoorten N., Van Impe K., Fedotov V., Dadali E., Auer-Grumbach M., Windpassinger C., Wagner K., Mitrovic Z., Hilton-Jones D., Talbot K., Martin J. J., Vasserman N., Tverskaya S., Polyakov A., Liem R. K., Gettemans J., Robberecht W., De Jonghe P., Timmerman V. (2004) Nat. Genet. 36, 602–606 - PubMed
-
- Houlden H., Laura M., Wavrant-De Vrièze F., Blake J., Wood N., Reilly M. M. (2008) Neurology 71, 1660–1668 - PubMed
-
- Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Nat. Struct. Mol. Biol. 12, 842–846 - PubMed
-
- Lelj-Garolla B., Mauk A. G. (2006) J. Biol. Chem. 281, 8169–8174 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

