Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development
- PMID: 20179138
- PMCID: PMC2845417
- DOI: 10.1105/tpc.109.072777
Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development
Abstract
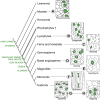
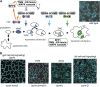
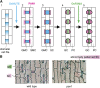
Stomata are microscopic valves on the plant epidermis that played a critical role in the evolution of land plants. Studies in the model dicot Arabidopsis thaliana have identified key transcription factors and signaling pathways controlling stomatal patterning and differentiation. Three paralogous Arabidopsis basic helix-loop-helix proteins, SPEECHLESS (SPCH), MUTE, and FAMA, mediate sequential steps of cell-state transitions together with their heterodimeric partners SCREAM (SCRM) and SCRM2. Cell-cell signaling components, including putative ligands, putative receptors, and mitogen-activated protein kinase cascades, orient asymmetric cell divisions and prevent overproduction and clustering of stomata. The recent availability of genome sequence and reverse genetics tools for model monocots and basal land plants allows for the examination of the conservation of genes important in stomatal patterning and differentiation. Studies in grasses have revealed that divergence of SPCH-MUTE-FAMA predates the evolutionary split of monocots and dicots and that these proteins show conserved and novel roles in stomatal differentiation. By contrast, specific asymmetric cell divisions in Arabidopsis and grasses require unique molecular components. Molecular phylogenetic analysis implies potential conservation of signaling pathways and prototypical functions of the transcription factors specifying stomatal differentiation.
Figures
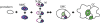

References
-
- Abrash E.B., Bergmann D.C. (2010). Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 - PubMed
-
- Apostolakos P., Panteris E., Galatis B. (1997). Microtubule and actin filament organization during stomatal morphogenesis in the fern Asplenium nidus. 1. Guard cell mother cell. Protoplasma 198: 93–106 - PubMed
-
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 - PubMed
-
- Bhave N.S., Veley K.M., Nadeau J.A., Lucas J.R., Bhave S.L., Sack F.D. (2009). TOO MANY MOUTHS promotes cell fate progression in stomatal development of Arabidopsis stems. Planta 229: 357–367 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

