Pulsed-field gel electrophoresis diversity of human and bovine clinical Salmonella isolates
- PMID: 20180633
- PMCID: PMC3132108
- DOI: 10.1089/fpd.2009.0424
Pulsed-field gel electrophoresis diversity of human and bovine clinical Salmonella isolates
Abstract
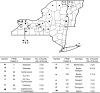
Pulsed-field gel electrophoresis (PFGE) characterization of 335 temporally and spatially matched clinical, bovine, and human Salmonella enterica subsp. enterica isolates revealed 167 XbaI PFGE patterns. These isolates were previously classified into 51 serotypes and 73 sequence types, as determined by multilocus sequence typing. Discriminatory power of PFGE (Simpson's index, D = 0.991) was considerably higher than that of multilocus sequence typing (D = 0.920) or serotyping (D = 0.913). Although 128 PFGE types each only represented a single isolate, 8 PFGE types represented >4 isolates, including (i) three serotype Enteritidis and Heidelberg patterns that were only identified among human isolates, (ii) two PFGE patterns (each representing serotypes Bardo and Newport) that were significantly more common among bovine isolates as compared with human isolates; (iii) two PFGE types that each includes two serotypes (4,5,12:i:- and Typhimurium; Thompson and 1,7:-:1,5); and (iv) one PFGE type that includes eight Typhimurium isolates from humans and cattle. Characterization of isolates collected over multiple farm visits indicated that given specific PFGE types persisted over time on 11 farms. On an additional seven farms, isolates with a given sequence type represented multiple PFGE type, which typically only differed by <3 bands, suggesting PFGE type diversification during strain persistence. Sixteen PFGE types were isolated from 2 or more farms, including two widely distributed serotype Newport-associated PFGE types each found on 10 farms. In six instances two or three human isolates collected in the same county in the same or consecutive months represented the same subtypes, suggesting small human case clusters. PFGE-based characterization and surveillance of human and animal isolates can provide improved understanding of Salmonella diversity and epidemiology, including identification of possible host-associated and common, widely distributed PFGE types.
Figures
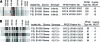

References
-
- Barrett TJ. Gerner-Smidt P. Swaminathan B. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog Dis. 2006;3:20–31. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

