Dealing with the problem of non-specific in situ mRNA hybridization signals associated with plant tissues undergoing programmed cell death
- PMID: 20181098
- PMCID: PMC2829549
- DOI: 10.1186/1746-4811-6-7
Dealing with the problem of non-specific in situ mRNA hybridization signals associated with plant tissues undergoing programmed cell death
Abstract
Background: In situ hybridization is a general molecular method typically used for the localization of mRNA transcripts in plants. The method provides a valuable tool to unravel the connection between gene expression and anatomy, especially in species such as pines which show large genome size and shortage of sequence information.
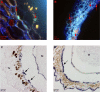
Results: In the present study, expression of the catalase gene (CAT) related to the scavenging of reactive oxygen species (ROS) and the polyamine metabolism related genes, diamine oxidase (DAO) and arginine decarboxylase (ADC), were localized in developing Scots pine (Pinus sylvestris L.) seeds. In addition to specific signals from target mRNAs, the probes continually hybridized non-specifically in the embryo surrounding region (ESR) of the megagametophyte tissue, in the remnants of the degenerated suspensors as well as in the cells of the nucellar layers, i.e. tissues exposed to cell death processes and extensive nucleic acid fragmentation during Scots pine seed development.
Conclusions: In plants, cell death is an integral part of both development and defence, and hence it is a common phenomenon in all stages of the life cycle. Our results suggest that extensive nucleic acid fragmentation during cell death processes can be a considerable source of non-specific signals in traditional in situ mRNA hybridization. Thus, the visualization of potential nucleic acid fragmentation simultaneously with the in situ mRNA hybridization assay may be necessary to ensure the correct interpretation of the signals in the case of non-specific hybridization of probes in plant tissues.
Figures


Similar articles
-
Expression of catalase and retinoblastoma-related protein genes associates with cell death processes in Scots pine zygotic embryogenesis.BMC Plant Biol. 2015 Mar 15;15:88. doi: 10.1186/s12870-015-0462-0. BMC Plant Biol. 2015. PMID: 25887788 Free PMC article.
-
Thermospermine Synthase (ACL5) and Diamine Oxidase (DAO) Expression Is Needed for Zygotic Embryogenesis and Vascular Development in Scots Pine.Front Plant Sci. 2019 Dec 20;10:1600. doi: 10.3389/fpls.2019.01600. eCollection 2019. Front Plant Sci. 2019. PMID: 31921249 Free PMC article.
-
Moderate stress responses and specific changes in polyamine metabolism characterize Scots pine somatic embryogenesis.Tree Physiol. 2016 Mar;36(3):392-402. doi: 10.1093/treephys/tpv136. Epub 2016 Jan 19. Tree Physiol. 2016. PMID: 26786537 Free PMC article.
-
Detection of mRNAs encoding peroxisomal proteins by non-radioactive in situ hybridization with digoxigenin-labelled cRNAs.Histochem Cell Biol. 1997 Oct-Nov;108(4-5):371-9. doi: 10.1007/s004180050178. Histochem Cell Biol. 1997. PMID: 9387930 Review.
-
Pine embryogenesis: many licences to kill for a new life.Plant Signal Behav. 2009 Oct;4(10):928-32. doi: 10.4161/psb.4.10.9535. Epub 2009 Oct 16. Plant Signal Behav. 2009. PMID: 19826239 Free PMC article. Review.
Cited by
-
Expression of catalase and retinoblastoma-related protein genes associates with cell death processes in Scots pine zygotic embryogenesis.BMC Plant Biol. 2015 Mar 15;15:88. doi: 10.1186/s12870-015-0462-0. BMC Plant Biol. 2015. PMID: 25887788 Free PMC article.
-
Visualization of spatial gene expression in plants by modified RNAscope fluorescent in situ hybridization.Plant Methods. 2020 May 15;16:71. doi: 10.1186/s13007-020-00614-4. eCollection 2020. Plant Methods. 2020. PMID: 32467719 Free PMC article.
-
Scots pine aminopropyltransferases shed new light on evolution of the polyamine biosynthesis pathway in seed plants.Ann Bot. 2018 May 11;121(6):1243-1256. doi: 10.1093/aob/mcy012. Ann Bot. 2018. PMID: 29462244 Free PMC article.
-
Thermospermine Synthase (ACL5) and Diamine Oxidase (DAO) Expression Is Needed for Zygotic Embryogenesis and Vascular Development in Scots Pine.Front Plant Sci. 2019 Dec 20;10:1600. doi: 10.3389/fpls.2019.01600. eCollection 2019. Front Plant Sci. 2019. PMID: 31921249 Free PMC article.
References
-
- Nitta H, Hicks DG, Skacel M, Pettay J, Grogan T, Tubbs RR. In: Molecular morphology in human tissues: techniques and applications. Hacker GW, Tubbs RR, editor. Boca Raton: CRC Press; 2004. High throughput morphological gene expression studies using automated mRNA in situ hybridization applications and tissue microarrays for post-genomic and clinical research; pp. 147–156.
LinkOut - more resources
Full Text Sources
Miscellaneous

