Correcting for intra-experiment variation in Illumina BeadChip data is necessary to generate robust gene-expression profiles
- PMID: 20181233
- PMCID: PMC2843619
- DOI: 10.1186/1471-2164-11-134
Correcting for intra-experiment variation in Illumina BeadChip data is necessary to generate robust gene-expression profiles
Abstract
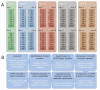
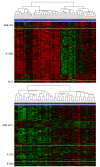
Background: Microarray technology is a popular means of producing whole genome transcriptional profiles, however high cost and scarcity of mRNA has led many studies to be conducted based on the analysis of single samples. We exploit the design of the Illumina platform, specifically multiple arrays on each chip, to evaluate intra-experiment technical variation using repeated hybridisations of universal human reference RNA (UHRR) and duplicate hybridisations of primary breast tumour samples from a clinical study.
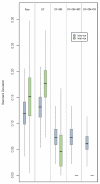
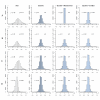
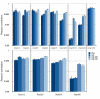
Results: A clear batch-specific bias was detected in the measured expressions of both the UHRR and clinical samples. This bias was found to persist following standard microarray normalisation techniques. However, when mean-centering or empirical Bayes batch-correction methods (ComBat) were applied to the data, inter-batch variation in the UHRR and clinical samples were greatly reduced. Correlation between replicate UHRR samples improved by two orders of magnitude following batch-correction using ComBat (ranging from 0.9833-0.9991 to 0.9997-0.9999) and increased the consistency of the gene-lists from the duplicate clinical samples, from 11.6% in quantile normalised data to 66.4% in batch-corrected data. The use of UHRR as an inter-batch calibrator provided a small additional benefit when used in conjunction with ComBat, further increasing the agreement between the two gene-lists, up to 74.1%.
Conclusion: In the interests of practicalities and cost, these results suggest that single samples can generate reliable data, but only after careful compensation for technical bias in the experiment. We recommend that investigators appreciate the propensity for such variation in the design stages of a microarray experiment and that the use of suitable correction methods become routine during the statistical analysis of the data.
Figures

References
-
- Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J Clin Oncol. 2002;20(7):1932–1941. - PubMed
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. - DOI - PubMed
-
- Baggerly KA, Coombes KR. Deriving Chemosensitivity from Cell Lines: Forensic Bioinformatics and Reproducible Research in High-Throughput Biology. Annals of Applied Statistics. in press .
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

