Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels
- PMID: 20181251
- PMCID: PMC2848037
- DOI: 10.1186/1471-2148-10-61
Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels
Abstract
Background: Although the overwhelming majority of genes found in angiosperms are members of gene families, and both gene- and genome-duplication are pervasive forces in plant genomes, some genes are sufficiently distinct from all other genes in a genome that they can be operationally defined as 'single copy'. Using the gene clustering algorithm MCL-tribe, we have identified a set of 959 single copy genes that are shared single copy genes in the genomes of Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera and Oryza sativa. To characterize these genes, we have performed a number of analyses examining GO annotations, coding sequence length, number of exons, number of domains, presence in distant lineages, such as Selaginella and Physcomitrella, and phylogenetic analysis to estimate copy number in other seed plants and to demonstrate their phylogenetic utility. We then provide examples of how these genes may be used in phylogenetic analyses to reconstruct organismal history, both by using extant coverage in EST databases for seed plants and de novo amplification via RT-PCR in the family Brassicaceae.
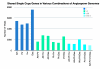
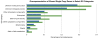
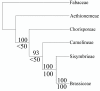
Results: There are 959 single copy nuclear genes shared in Arabidopsis, Populus, Vitis and Oryza ["APVO SSC genes"]. The majority of these genes are also present in the Selaginella and Physcomitrella genomes. Public EST sets for 197 species suggest that most of these genes are present across a diverse collection of seed plants, and appear to exist as single or very low copy genes, though exceptions are seen in recently polyploid taxa and in lineages where there is significant evidence for a shared large-scale duplication event. Genes encoding proteins localized in organelles are more commonly single copy than expected by chance, but the evolutionary forces responsible for this bias are unknown.Regardless of the evolutionary mechanisms responsible for the large number of shared single copy genes in diverse flowering plant lineages, these genes are valuable for phylogenetic and comparative analyses. Eighteen of the APVO SSC single copy genes were amplified in the Brassicaceae using RT-PCR and directly sequenced. Alignments of these sequences provide improved resolution of Brassicaceae phylogeny compared to recent studies using plastid and ITS sequences. An analysis of sequences from 13 APVO SSC genes from 69 species of seed plants, derived mainly from public EST databases, yielded a phylogeny that was largely congruent with prior hypotheses based on multiple plastid sequences. Whereas single gene phylogenies that rely on EST sequences have limited bootstrap support as the result of limited sequence information, concatenated alignments result in phylogenetic trees with strong bootstrap support for already established relationships. Overall, these single copy nuclear genes are promising markers for phylogenetics, and contain a greater proportion of phylogenetically-informative sites than commonly used protein-coding sequences from the plastid or mitochondrial genomes.
Conclusions: Putatively orthologous, shared single copy nuclear genes provide a vast source of new evidence for plant phylogenetics, genome mapping, and other applications, as well as a substantial class of genes for which functional characterization is needed. Preliminary evidence indicates that many of the shared single copy nuclear genes identified in this study may be well suited as markers for addressing phylogenetic hypotheses at a variety of taxonomic levels.
Figures

Similar articles
-
Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice.Biol Direct. 2015 Sep 8;10:48. doi: 10.1186/s13062-015-0076-3. Biol Direct. 2015. PMID: 26350041 Free PMC article.
-
Analyses of the oligopeptide transporter gene family in poplar and grape.BMC Genomics. 2011 Sep 26;12:465. doi: 10.1186/1471-2164-12-465. BMC Genomics. 2011. PMID: 21943393 Free PMC article.
-
Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors.Ann Bot. 2011 May;107(7):1141-57. doi: 10.1093/aob/mcr034. Epub 2011 Mar 7. Ann Bot. 2011. PMID: 21385782 Free PMC article.
-
Utility of low-copy nuclear gene sequences in plant phylogenetics.Crit Rev Biochem Mol Biol. 2002;37(3):121-47. doi: 10.1080/10409230290771474. Crit Rev Biochem Mol Biol. 2002. PMID: 12139440 Review.
-
Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs.J Plant Res. 2006 Jan;119(1):11-21. doi: 10.1007/s10265-005-0253-z. Epub 2006 Jan 13. J Plant Res. 2006. PMID: 16411016 Review.
Cited by
-
Genome-wide signatures of plastid-nuclear coevolution point to repeated perturbations of plastid proteostasis systems across angiosperms.Plant Cell. 2021 May 31;33(4):980-997. doi: 10.1093/plcell/koab021. Plant Cell. 2021. PMID: 33764472 Free PMC article.
-
Phylotranscriptomic Analyses of Mycoheterotrophic Monocots Show a Continuum of Convergent Evolutionary Changes in Expressed Nuclear Genes From Three Independent Nonphotosynthetic Lineages.Genome Biol Evol. 2023 Jan 4;15(1):evac183. doi: 10.1093/gbe/evac183. Genome Biol Evol. 2023. PMID: 36582124 Free PMC article.
-
Gene prediction and annotation in Penstemon (Plantaginaceae): A workflow for marker development from extremely low-coverage genome sequencing.Appl Plant Sci. 2014 Dec 4;2(12):apps.1400044. doi: 10.3732/apps.1400044. eCollection 2014 Dec. Appl Plant Sci. 2014. PMID: 25506519 Free PMC article.
-
POInTbrowse: orthology prediction and synteny exploration for paleopolyploid genomes.BMC Bioinformatics. 2023 Apr 27;24(1):174. doi: 10.1186/s12859-023-05298-w. BMC Bioinformatics. 2023. PMID: 37106333 Free PMC article.
-
Polyploidy and diploidization in soybean.Mol Breed. 2023 Jun 6;43(6):51. doi: 10.1007/s11032-023-01396-y. eCollection 2023 Jun. Mol Breed. 2023. PMID: 37313224 Free PMC article.
References
-
- Small RL, Cronn RC, Wendel JF. Use of nuclear genes for phylogeny reconstruction in plants. Australian Systematic Botany. 2004;17:145–170. doi: 10.1071/SB03015. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

