Avian reovirus microNS protein forms homo-oligomeric inclusions in a microtubule-independent fashion, which involves specific regions of its C-terminal domain
- PMID: 20181708
- PMCID: PMC2863718
- DOI: 10.1128/JVI.02534-09
Avian reovirus microNS protein forms homo-oligomeric inclusions in a microtubule-independent fashion, which involves specific regions of its C-terminal domain
Abstract
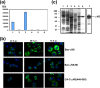
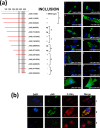
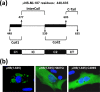
Members of the genus Orthoreovirus replicate in cytoplasmic inclusions termed viral factories. Compelling evidence suggests that the nonstructural protein microNS forms the matrix of the factories and recruits specific viral proteins to these structures. In the first part of this study, we analyzed the properties of avian reovirus factories and microNS-derived inclusions and found that they are nonaggresome cytoplasmic globular structures not associated with the cytoskeleton which do not require an intact microtubule network for formation and maturation. We next investigated the capacity of avian reovirus microNS to form inclusions in transfected and baculovirus-infected cells. Our results showed that microNS is the main component of the inclusions formed by recombinant baculovirus expression. This, and the fact that microNS is able to self-associate inside the cell, suggests that microNS monomers contain all the interacting domains required for inclusion formation. Examination of the inclusion-forming capacities of truncated microNS versions allowed us to identify the region spanning residues 448 to 635 of microNS as the smallest that was inclusion competent, although residues within the region 140 to 380 seem to be involved in inclusion maturation. Finally, we investigated the roles that four different motifs present in microNS(448-635) play in inclusion formation, and the results suggest that the C-terminal tail domain is a key determinant in dictating the initial orientation of monomer-to-monomer contacts to form basal oligomers that control inclusion shape and inclusion-forming efficiency. Our results contribute to an understanding of the generation of structured protein aggregates that escape the cellular mechanisms of protein recycling.
Figures





References
-
- Attoui, H., F. Billoir, P. Biagini, P. de Micco, and X. de Lamballerie. 2000. Complete sequence determination and genetic analysis of Banna virus and Kadipiro virus: proposal for assignment to a new genus (Seadornavirus) within the family Reoviridae. J. Gen. Virol. 81:1507-1515. - PubMed
-
- Bodelon, G., L. Labrada, J. Martinez-Costas, and J. Benavente. 2001. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology 290:181-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

