Structural basis of dimerization-dependent ubiquitination by the SCF(Fbx4) ubiquitin ligase
- PMID: 20181953
- PMCID: PMC2859552
- DOI: 10.1074/jbc.M110.111518
Structural basis of dimerization-dependent ubiquitination by the SCF(Fbx4) ubiquitin ligase
Abstract
The F-box proteins are the substrate recognition subunits of the SCF (Skp1-Cul1-Rbx1-F- box protein) ubiquitin ligase complexes that control the stability of numerous regulators in eukaryotic cells. Here we show that dimerization of the F-box protein Fbx4 is essential for SCF(Fbx4) (the superscript denotes the F-box protein) ubiquitination activity toward the telomere regulator Pin2 (also known as TRF1). The crystal structure of Fbx4 in complex with an adaptor protein Skp1 reveals an antiparallel dimer configuration in which the linker domain of Fbx4 interacts with the C-terminal substrate-binding domain of the other protomer, whereas the C-terminal domain of the protein adopts a compact alpha/beta fold distinct from those of known F-box proteins. Biochemical studies indicate that both the N-terminal domain and a loop connecting the linker and C-terminal domain of Fbx4 are critical for the dimerization and activation of the protein. Our findings provide a framework for understanding the role of F-box dimerization in the SCF-mediated ubiquitination reaction.
Figures


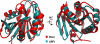

Similar articles
-
Structural basis of selective ubiquitination of TRF1 by SCFFbx4.Dev Cell. 2010 Feb 16;18(2):214-25. doi: 10.1016/j.devcel.2010.01.007. Dev Cell. 2010. PMID: 20159592 Free PMC article.
-
Phosphorylation-dependent regulation of SCF(Fbx4) dimerization and activity involves a novel component, 14-3-3ɛ.Oncogene. 2011 Apr 28;30(17):1995-2002. doi: 10.1038/onc.2010.584. Epub 2011 Jan 17. Oncogene. 2011. PMID: 21242966 Free PMC article.
-
Structure of the CUL1-RBX1-SKP1-FBXO4 SCF ubiquitin ligase complex.Biochem Biophys Res Commun. 2024 Nov 26;735:150811. doi: 10.1016/j.bbrc.2024.150811. Epub 2024 Oct 11. Biochem Biophys Res Commun. 2024. PMID: 39406020
-
The SCF ubiquitin ligase: insights into a molecular machine.Nat Rev Mol Cell Biol. 2004 Sep;5(9):739-51. doi: 10.1038/nrm1471. Nat Rev Mol Cell Biol. 2004. PMID: 15340381 Review.
-
Structure and Function of Fbxo7/PARK15 in Parkinson's Disease.Curr Protein Pept Sci. 2017;18(7):715-724. doi: 10.2174/1389203717666160311121433. Curr Protein Pept Sci. 2017. PMID: 26965690 Review.
Cited by
-
Arabidopsis Tubby domain-containing F-box proteins positively regulate immunity by modulating PI4Kβ protein levels.New Phytol. 2023 Oct;240(1):354-371. doi: 10.1111/nph.19187. Epub 2023 Aug 12. New Phytol. 2023. PMID: 37571862 Free PMC article.
-
CUL3 E3 ligases in plant development and environmental response.Nat Plants. 2021 Jan;7(1):6-16. doi: 10.1038/s41477-020-00833-6. Epub 2021 Jan 15. Nat Plants. 2021. PMID: 33452490 Free PMC article. Review.
-
The E3 Ubiquitin Ligase Fbxo4 Functions as a Tumor Suppressor: Its Biological Importance and Therapeutic Perspectives.Cancers (Basel). 2022 Apr 25;14(9):2133. doi: 10.3390/cancers14092133. Cancers (Basel). 2022. PMID: 35565262 Free PMC article. Review.
-
Regenerative approaches to preserve pancreatic β-cell mass and function in diabetes pathogenesis.Endocrine. 2022 Feb;75(2):338-350. doi: 10.1007/s12020-021-02941-5. Epub 2021 Nov 25. Endocrine. 2022. PMID: 34825343 Review.
-
Functional coupling of TRPM2 and extrasynaptic NMDARs exacerbates excitotoxicity in ischemic brain injury.Neuron. 2022 Jun 15;110(12):1944-1958.e8. doi: 10.1016/j.neuron.2022.03.021. Epub 2022 Apr 13. Neuron. 2022. PMID: 35421327 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

