Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein
- PMID: 20181956
- PMCID: PMC2859521
- DOI: 10.1074/jbc.M109.089540
Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein
Abstract
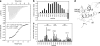
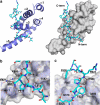
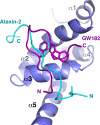
Poly(A)-binding protein (PABPC1) is involved in multiple aspects of mRNA processing and translation. It is a component of RNA stress granules and binds the RNA-induced silencing complex to promote degradation of silenced mRNAs. Here, we report the crystal structures of the C-terminal Mlle (or PABC) domain in complex with peptides from GW182 (TNRC6C) and Ataxin-2. The structures reveal overlapping binding sites but with unexpected diversity in the peptide conformation and residues involved in binding. The mutagenesis and binding studies show low to submicromolar binding affinity with overlapping but distinct specificity determinants. These results rationalize the role of the Mlle domain of PABPC1 in microRNA-mediated mRNA deadenylation and suggest a more general function in the assembly of cytoplasmic RNA granules.
Figures
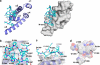
References
-
- Kühn U., Wahle E. (2004) Biochim. Biophys. Acta 1678, 67–84 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

