Approaching cellular and molecular resolution of auxin biosynthesis and metabolism
- PMID: 20182605
- PMCID: PMC2827909
- DOI: 10.1101/cshperspect.a001594
Approaching cellular and molecular resolution of auxin biosynthesis and metabolism
Abstract
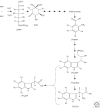
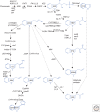
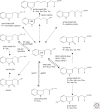
There is abundant evidence of multiple biosynthesis pathways for the major naturally occurring auxin in plants, indole-3-acetic acid (IAA), and examples of differential use of two general routes of IAA synthesis, namely Trp-dependent and Trp-independent. Although none of these pathways has been completely defined, we now have examples of specific IAA biosynthetic pathways playing a role in developmental processes by way of localized IAA synthesis, causing us to rethink the interactions between IAA synthesis, transport, and signaling. Recent work also points to some IAA biosynthesis pathways being specific to families within the plant kingdom, whereas others appear to be more ubiquitous. An important advance within the past 5 years is our ability to monitor IAA biosynthesis and metabolism at increasingly higher resolution.
Figures
Similar articles
-
Auxin: regulation, action, and interaction.Ann Bot. 2005 Apr;95(5):707-35. doi: 10.1093/aob/mci083. Epub 2005 Mar 4. Ann Bot. 2005. PMID: 15749753 Free PMC article. Review.
-
Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants.Mol Plant. 2012 Mar;5(2):334-8. doi: 10.1093/mp/ssr104. Epub 2011 Dec 8. Mol Plant. 2012. PMID: 22155950 Free PMC article. Review.
-
Auxin and Tryptophan Homeostasis Are Facilitated by the ISS1/VAS1 Aromatic Aminotransferase in Arabidopsis.Genetics. 2015 Sep;201(1):185-99. doi: 10.1534/genetics.115.180356. Epub 2015 Jul 10. Genetics. 2015. PMID: 26163189 Free PMC article.
-
Current aspects of auxin biosynthesis in plants.Biosci Biotechnol Biochem. 2016;80(1):34-42. doi: 10.1080/09168451.2015.1086259. Epub 2015 Sep 12. Biosci Biotechnol Biochem. 2016. PMID: 26364770 Review.
-
Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis.Plant J. 2014 Feb;77(3):352-66. doi: 10.1111/tpj.12399. Epub 2014 Jan 16. Plant J. 2014. PMID: 24299123
Cited by
-
New Insights into the TIFY Gene Family of Brassica napus and Its Involvement in the Regulation of Shoot Branching.Int J Mol Sci. 2023 Dec 4;24(23):17114. doi: 10.3390/ijms242317114. Int J Mol Sci. 2023. PMID: 38069438 Free PMC article.
-
Indole-3-acetic acid is a physiological inhibitor of TORC1 in yeast.PLoS Genet. 2021 Mar 9;17(3):e1009414. doi: 10.1371/journal.pgen.1009414. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33690632 Free PMC article.
-
Quantification of reaction cycle parameters for an essential molecular switch in an auxin-responsive transcription circuit in rice.Proc Natl Acad Sci U S A. 2019 Feb 12;116(7):2589-2594. doi: 10.1073/pnas.1817038116. Epub 2019 Jan 29. Proc Natl Acad Sci U S A. 2019. PMID: 30696765 Free PMC article.
-
Metabolic profiles of 2-oxindole-3-acetyl-amino acid conjugates differ in various plant species.Front Plant Sci. 2023 Jul 18;14:1217421. doi: 10.3389/fpls.2023.1217421. eCollection 2023. Front Plant Sci. 2023. PMID: 37534287 Free PMC article.
-
Biosynthetic Pathways of Hormones in Plants.Metabolites. 2023 Jul 25;13(8):884. doi: 10.3390/metabo13080884. Metabolites. 2023. PMID: 37623827 Free PMC article. Review.
References
-
- Bartel B, Fink G 1995. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268:1745–1748 - PubMed
-
- Barth C, Jander G 2006. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. The Plant Journal 46:549–562 - PubMed
-
- Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al.2009. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
