Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens
- PMID: 20184753
- PMCID: PMC2838877
- DOI: 10.1186/1471-2180-10-64
Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens
Abstract
Background: Intracellular bacterial pathogens depend on acquisition of iron for their success as pathogens. The host cell requires iron as an essential component for cellular functions that include innate immune defense mechanisms. The transferrin receptor TfR1 plays an important part for delivering iron to the host cell during infection. Its expression can be modulated by infection, but its essentiality for bacterial intracellular survival has not been directly investigated.
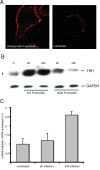
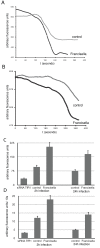
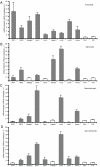
Results: We identified two distinct iron-handling scenarios for two different bacterial pathogens. Francisella tularensis drives an active iron acquisition program via the TfR1 pathway program with induction of ferrireductase (Steap3), iron membrane transporter Dmt1, and iron regulatory proteins IRP1 and IRP2, which is associated with a sustained increase of the labile iron pool inside the macrophage. Expression of TfR1 is critical for Francisella's intracellular proliferation. This contrasts with infection of macrophages by wild-type Salmonella typhimurium, which does not require expression of TfR1 for successful intracellular survival. Macrophages infected with Salmonella lack significant induction of Dmt1, Steap3, and IRP1, and maintain their labile iron pool at normal levels.
Conclusion: The distinction between two different phenotypes of iron utilization by intracellular pathogens will allow further characterization and understanding of host-cell iron metabolism and its modulation by intracellular bacteria.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

