Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans
- PMID: 20184761
- PMCID: PMC2841656
- DOI: 10.1186/1471-2148-10-62
Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans
Abstract
Background: Crustacean Hyperglycemic Hormone (CHH) family peptides are neurohormones known to regulate several important functions in decapod crustaceans such as ionic and energetic metabolism, molting and reproduction. The structural conservation of these peptides, together with the variety of functions they display, led us to investigate their evolutionary history. CHH family peptides exist in insects (Ion Transport Peptides) and may be present in all ecdysozoans as well. In order to extend the evolutionary study to the entire family, CHH family peptides were thus searched in taxa outside decapods, where they have been, to date, poorly investigated.
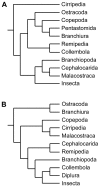
Results: CHH family peptides were characterized by molecular cloning in a branchiopod crustacean, Daphnia magna, and in a collembolan, Folsomia candida. Genes encoding such peptides were also rebuilt in silico from genomic sequences of another branchiopod, a chelicerate and two nematodes. These sequences were included in updated datasets to build phylogenies of the CHH family in pancrustaceans. These phylogenies suggest that peptides found in Branchiopoda and Collembola are more closely related to insect ITPs than to crustacean CHHs. Datasets were also used to support a phylogenetic hypothesis about pancrustacean relationships, which, in addition to gene structures, allowed us to propose two evolutionary scenarios of this multigenic family in ecdysozoans.
Conclusions: Evolutionary scenarios suggest that CHH family genes of ecdysozoans originate from an ancestral two-exon gene, and genes of arthropods from a three-exon one. In malacostracans, the evolution of the CHH family has involved several duplication, insertion or deletion events, leading to neuropeptides with a wide variety of functions, as observed in decapods. This family could thus constitute a promising model to investigate the links between gene duplications and functional divergence.
Figures



Similar articles
-
The Crustacean Hyperglycemic Hormone Superfamily: Progress Made in the Past Decade.Front Endocrinol (Lausanne). 2020 Oct 1;11:578958. doi: 10.3389/fendo.2020.578958. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33117290 Free PMC article. Review.
-
In silico analysis of crustacean hyperglycemic hormone family.Mar Biotechnol (NY). 2005 May-Jun;7(3):193-206. doi: 10.1007/s10126-004-0020-5. Epub 2005 Jun 8. Mar Biotechnol (NY). 2005. PMID: 15933902
-
Comparative genomic analysis of crustacean hyperglycemic hormone (CHH) neuropeptide genes across diverse crustacean species.F1000Res. 2018 Jan 23;7:100. doi: 10.12688/f1000research.13732.1. eCollection 2018. F1000Res. 2018. PMID: 30356453 Free PMC article.
-
Two type I crustacean hyperglycemic hormone (CHH) genes in Morotoge shrimp (Pandalopsis japonica): cloning and expression of eyestalk and pericardial organ isoforms produced by alternative splicing and a novel type I CHH with predicted structure shared with type II CHH peptides.Comp Biochem Physiol B Biochem Mol Biol. 2012 Aug;162(4):88-99. doi: 10.1016/j.cbpb.2012.04.003. Epub 2012 Apr 13. Comp Biochem Physiol B Biochem Mol Biol. 2012. PMID: 22525298
-
Crustacean neuropeptide genes of the CHH/MIH/GIH family: implications from molecular studies.Gen Comp Endocrinol. 2003 Dec;134(3):214-9. doi: 10.1016/s0016-6480(03)00263-6. Gen Comp Endocrinol. 2003. PMID: 14636627 Review.
Cited by
-
Function-driven discovery of neuropeptides with mass spectrometry-based tools.Protein Pept Lett. 2013 Jun;20(6):681-94. doi: 10.2174/0929866511320060007. Protein Pept Lett. 2013. PMID: 22630128 Free PMC article. Review.
-
Manual classification strategies in the ECOD database.Proteins. 2015 Jul;83(7):1238-51. doi: 10.1002/prot.24818. Epub 2015 May 8. Proteins. 2015. PMID: 25917548 Free PMC article.
-
A mass spectrometry-based method to screen for α-amidated peptides.Proteomics. 2012 Jan;12(2):173-82. doi: 10.1002/pmic.201100327. Epub 2011 Dec 14. Proteomics. 2012. PMID: 22106059 Free PMC article.
-
Differential effects of silencing crustacean hyperglycemic hormone gene expression on the metabolic profiles of the muscle and hepatopancreas in the crayfish Procambarus clarkii.PLoS One. 2017 Feb 16;12(2):e0172557. doi: 10.1371/journal.pone.0172557. eCollection 2017. PLoS One. 2017. PMID: 28207859 Free PMC article.
-
Genome-Wide Identification and Expression of Neuropeptides and Their Expression Patterns After RNAi of CHH Genes in Pacific White Shrimp Litopenaeus vannamei.Biology (Basel). 2024 Dec 11;13(12):1038. doi: 10.3390/biology13121038. Biology (Basel). 2024. PMID: 39765705 Free PMC article.
References
-
- Böcking D, Dircksen H, Keller R. In: The Crustacean Nervous System. Wiese K, editor. Berlin, Heidelberg, New York: Springer; 2002. The crustacean neuropeptides of the CHH/MIH/GIH family: structures and biological activities; pp. 84–97.
-
- Soyez D. In: Recent Advances in Marine Biotechnology. Fingerman M, Nagabhushanam R, editor. Vol. 10. Plymouth, U.K.: Science Publishers; 2003. Recent data on the crustacean hyperglycemic hormone family; pp. 279–301.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

