A population genetic hidden Markov model for detecting genomic regions under selection
- PMID: 20185453
- PMCID: PMC2912474
- DOI: 10.1093/molbev/msq053
A population genetic hidden Markov model for detecting genomic regions under selection
Abstract
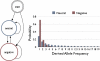
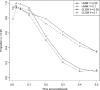
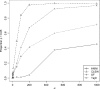
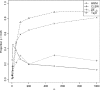
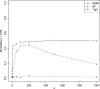
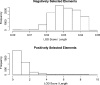
Recently, hidden Markov models have been applied to numerous problems in genomics. Here, we introduce an explicit population genetics hidden Markov model (popGenHMM) that uses single nucleotide polymorphism (SNP) frequency data to identify genomic regions that have experienced recent selection. Our popGenHMM assumes that SNP frequencies are emitted independently following diffusion approximation expectations but that neighboring SNP frequencies are partially correlated by selective state. We give results from the training and application of our popGenHMM to a set of early release data from the Drosophila Population Genomics Project (dpgp.org) that consists of approximately 7.8 Mb of resequencing from 32 North American Drosophila melanogaster lines. These results demonstrate the potential utility of our model, making predictions based on the site frequency spectrum (SFS) for regions of the genome that represent selected elements.
Figures



Similar articles
-
Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations.Genetics. 2010 Jul;185(3):907-22. doi: 10.1534/genetics.110.116459. Epub 2010 Apr 20. Genetics. 2010. PMID: 20407129 Free PMC article.
-
Detecting selective sweeps: a new approach based on hidden markov models.Genetics. 2009 Apr;181(4):1567-78. doi: 10.1534/genetics.108.100032. Epub 2009 Feb 9. Genetics. 2009. PMID: 19204373 Free PMC article.
-
Detecting highly differentiated copy-number variants from pooled population sequencing.Pac Symp Biocomput. 2013:344-55. Pac Symp Biocomput. 2013. PMID: 23424139 Free PMC article.
-
Population genomics of transposable elements in Drosophila.Annu Rev Genet. 2014;48:561-81. doi: 10.1146/annurev-genet-120213-092359. Epub 2014 Oct 1. Annu Rev Genet. 2014. PMID: 25292358 Review.
-
A population genomic approach to map recent positive selection in model species.Mol Ecol. 2008 Aug;17(16):3585-98. doi: 10.1111/j.1365-294X.2008.03852.x. Mol Ecol. 2008. PMID: 18627454 Review.
Cited by
-
Evolutionary forces shaping genomic islands of population differentiation in humans.BMC Genomics. 2012 Mar 22;13:107. doi: 10.1186/1471-2164-13-107. BMC Genomics. 2012. PMID: 22439654 Free PMC article.
-
Supervised Machine Learning for Population Genetics: A New Paradigm.Trends Genet. 2018 Apr;34(4):301-312. doi: 10.1016/j.tig.2017.12.005. Epub 2018 Jan 10. Trends Genet. 2018. PMID: 29331490 Free PMC article. Review.
-
Detecting Selection from Linked Sites Using an F-Model.Genetics. 2020 Dec;216(4):1205-1215. doi: 10.1534/genetics.120.303780. Epub 2020 Oct 16. Genetics. 2020. PMID: 33067324 Free PMC article.
-
Genomics of isolation in hybrids.Philos Trans R Soc Lond B Biol Sci. 2012 Feb 5;367(1587):439-50. doi: 10.1098/rstb.2011.0196. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22201173 Free PMC article.
-
A population genetics-phylogenetics approach to inferring natural selection in coding sequences.PLoS Genet. 2011 Dec;7(12):e1002395. doi: 10.1371/journal.pgen.1002395. Epub 2011 Dec 1. PLoS Genet. 2011. PMID: 22144911 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

