OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation
- PMID: 20185555
- PMCID: PMC2865371
- DOI: 10.1093/hmg/ddq088
OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation
Abstract
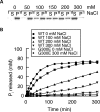
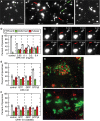
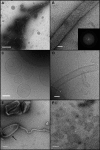
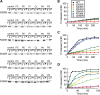
The dynamin-related GTPase OPA1 is mutated in autosomal dominant optic atrophy (DOA) (Kjer type), an inherited neuropathy of the retinal ganglion cells. OPA1 is essential for the fusion of the inner mitochondrial membranes, but its mechanism of action remains poorly understood. Here we show that OPA1 has a low basal rate of GTP hydrolysis that is dramatically enhanced by association with liposomes containing negative phospholipids such as cardiolipin. Lipid association triggers assembly of OPA1 into higher order oligomers. In addition, we find that OPA1 can promote the protrusion of lipid tubules from the surface of cardiolipin-containing liposomes. In such lipid protrusions, OPA1 assemblies are observed on the outside of the lipid tubule surface, a protein-membrane topology similar to that of classical dynamins. The membrane tubulation activity of OPA1 is suppressed by GTPgammaS. OPA1 disease alleles associated with DOA display selective defects in several activities, including cardiolipin association, GTP hydrolysis and membrane tubulation. These findings indicate that interaction of OPA1 with membranes can stimulate higher order assembly, enhance GTP hydrolysis and lead to membrane deformation into tubules.
Figures



References
-
- Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi:10.1038/nrm2275. - DOI - PubMed
-
- Song Z., Ghochani M., McCaffery J.M., Frey T.G., Chan D.C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell. 2009;20:3525–3532. doi:10.1091/mbc.E09-03-0252. - DOI - PMC - PubMed
-
- Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A.S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi:10.1074/jbc.M211311200. - DOI - PubMed
-
- Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi:10.1083/jcb.200704110. - DOI - PMC - PubMed
-
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi:10.1016/j.cell.2006.09.021. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

