Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method
- PMID: 20185571
- PMCID: PMC2879535
- DOI: 10.1093/nar/gkq107
Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method
Abstract
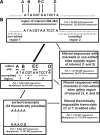
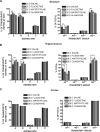
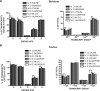
RNA editing is a post-transcriptional modification of pre-mRNA that results in increased diversity in transcriptomes and proteomes. It occurs in a wide variety of eukaryotic organisms and in some viruses. One of the most common forms of pre-mRNA editing is A-to-I editing, in which adenosine is deaminated to inosine, which is read as guanosine during translation. This phenomenon has been observed in numerous transcripts, including the mammalian 5-HT(2C) receptor, which can be edited at five distinct sites. Methods used to date to quantify 5-HT(2C) receptor editing are labor-intensive, expensive and provide limited information regarding the relative abundance of 5-HT(2C) receptor editing variants. Here, we present a novel, ultra high-throughput method to quantify 5-HT(2C) receptor editing, compare it to a more conventional method, and use it to assess the effect of a range of genetic and pharmacologic manipulations on 5-HT(2C) editing. We conclude that this new method is powerful and economical, and we provide evidence that alterations in 5-HT(2C) editing appear to be a result of regional changes in brain activity, rather than a mechanism to normalize 5-HT(2C) signaling.
Figures
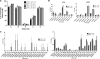
References
-
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. - PubMed
-
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. - PubMed
-
- Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. - PubMed
-
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

