Transcriptional regulation of the base excision repair pathway by BRCA1
- PMID: 20185827
- PMCID: PMC2885188
- DOI: 10.1074/jbc.M110.104430
Transcriptional regulation of the base excision repair pathway by BRCA1
Abstract
Inactivation of the breast cancer susceptibility gene BRCA1 plays a significant role in the development of a subset of breast cancers, although the major tumor suppressor function of this gene remains unclear. Previously, we showed that BRCA1 induces antioxidant-response gene expression and protects cells against oxidative stress. We now report that BRCA1 stimulates the base excision repair pathway, a major mechanism for the repair of oxidized DNA, by stimulating the activity of key base excision repair (BER) enzymes, including 8-oxoguanine DNA glycosylase (OGG1), the DNA glycosylase NTH1, and the apurinic endonuclease redox factor 1/apurinic endonuclease 1 (REF1/APE1), in human breast carcinoma cells. The increase in BER enzyme activity appears to be due, primarily, to an increase in enzyme expression. The ability of BRCA1 to stimulate the expression of the three BER enzymes and to enhance NTH1 promoter activity was dependent upon the octamer-binding transcription factor OCT1. Finally, we found that OGG1, NTH1, and REF1/APE1 each contribute to the BRCA1 protection against oxidative stress due to hydrogen peroxide and that hydrogen peroxide stimulates the expression of BRCA1 and the three BER enzymes. These findings identify a novel mechanism through which BRCA1 may regulate the repair of oxidative DNA damage.
Figures





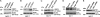

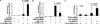

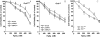

Similar articles
-
The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation.DNA Repair (Amst). 2018 Apr;64:10-25. doi: 10.1016/j.dnarep.2018.02.001. Epub 2018 Feb 11. DNA Repair (Amst). 2018. PMID: 29475157
-
Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress.Neuroscience. 2007 Apr 14;145(4):1201-12. doi: 10.1016/j.neuroscience.2006.10.010. Epub 2006 Nov 13. Neuroscience. 2007. PMID: 17101234
-
TRIM26 Maintains Cell Survival in Response to Oxidative Stress through Regulating DNA Glycosylase Stability.Int J Mol Sci. 2022 Oct 1;23(19):11613. doi: 10.3390/ijms231911613. Int J Mol Sci. 2022. PMID: 36232914 Free PMC article.
-
Transcription coupled base excision repair in mammalian cells: So little is known and so much to uncover.DNA Repair (Amst). 2021 Nov;107:103204. doi: 10.1016/j.dnarep.2021.103204. Epub 2021 Aug 6. DNA Repair (Amst). 2021. PMID: 34390916 Review.
-
Base excision repair modulation as a risk factor for human cancers.Mol Aspects Med. 2007 Jun-Aug;28(3-4):258-75. doi: 10.1016/j.mam.2007.05.003. Epub 2007 Jun 2. Mol Aspects Med. 2007. PMID: 17628657 Review.
Cited by
-
An Excimer Clamp for Measuring Damaged-Base Excision by the DNA Repair Enzyme NTH1.Angew Chem Int Ed Engl. 2020 May 4;59(19):7450-7455. doi: 10.1002/anie.202001516. Epub 2020 Mar 17. Angew Chem Int Ed Engl. 2020. PMID: 32109332 Free PMC article.
-
DIM (3,3'-diindolylmethane) confers protection against ionizing radiation by a unique mechanism.Proc Natl Acad Sci U S A. 2013 Nov 12;110(46):18650-5. doi: 10.1073/pnas.1308206110. Epub 2013 Oct 14. Proc Natl Acad Sci U S A. 2013. PMID: 24127581 Free PMC article.
-
XRCC1 deficient triple negative breast cancers are sensitive to ATR, ATM and Wee1 inhibitor either alone or in combination with olaparib.Ther Adv Med Oncol. 2020 Dec 21;12:1758835920974201. doi: 10.1177/1758835920974201. eCollection 2020. Ther Adv Med Oncol. 2020. PMID: 33425022 Free PMC article.
-
Regulation of DNA glycosylases and their role in limiting disease.Free Radic Res. 2012 Apr;46(4):460-78. doi: 10.3109/10715762.2012.655730. Epub 2012 Feb 6. Free Radic Res. 2012. PMID: 22300253 Free PMC article. Review.
-
DNA repair prognostic index modelling reveals an essential role for base excision repair in influencing clinical outcomes in ER negative and triple negative breast cancers.Oncotarget. 2015 Sep 8;6(26):21964-78. doi: 10.18632/oncotarget.4157. Oncotarget. 2015. PMID: 26267318 Free PMC article.
References
-
- D'Errico M., Parlanti E., Dogliotti E. (2008) Mutat. Res. 659, 4–14 - PubMed
-
- Mitra S., Hazra T. K., Roy R., Ikeda S., Biswas T., Lock J., Boldogh I., Izumi T. (1997) Mol. Cells 7, 305–312 - PubMed
-
- Izumi T., Wiederhold L. R., Roy G., Roy R., Jaiswal A., Bhakat K. K., Mitra S., Hazra T. K. (2003) Toxicology 193, 43–65 - PubMed
-
- Kelley M. R., Parsons S. H. (2001) Antioxid. Redox. Signal. 3, 671–683 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

