A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome
- PMID: 20186120
- PMCID: PMC2845271
- DOI: 10.1038/emboj.2010.14
A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome
Abstract
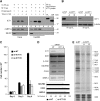
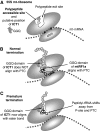
Bioinformatic analysis classifies the human protein encoded by immature colon carcinoma transcript-1 (ICT1) as one of a family of four putative mitochondrial translation release factors. However, this has not been supported by any experimental evidence. As only a single member of this family, mtRF1a, is required to terminate the synthesis of all 13 mitochondrially encoded polypeptides, the true physiological function of ICT1 was unclear. Here, we report that ICT1 is an essential mitochondrial protein, but unlike the other family members that are matrix-soluble, ICT1 has become an integral component of the human mitoribosome. Release-factor assays show that although ICT1 has retained its ribosome-dependent PTH activity, this is codon-independent; consistent with its loss of both domains that promote codon recognition in class-I release factors. Mutation of the GGQ domain common to ribosome-dependent PTHs causes a loss of activity in vitro and, crucially, a loss of cell viability, in vivo. We suggest that ICT1 may be essential for hydrolysis of prematurely terminated peptidyl-tRNA moieties in stalled mitoribosomes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
ICT1 comes to the rescue of mitochondrial ribosomes.EMBO J. 2010 Mar 17;29(6):1019-20. doi: 10.1038/emboj.2010.22. EMBO J. 2010. PMID: 20234387 Free PMC article. No abstract available.
Similar articles
-
Ribosome rescue and translation termination at non-standard stop codons by ICT1 in mammalian mitochondria.PLoS Genet. 2014 Sep 18;10(9):e1004616. doi: 10.1371/journal.pgen.1004616. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25233460 Free PMC article.
-
Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality.J Mol Biol. 2010 Nov 26;404(2):260-73. doi: 10.1016/j.jmb.2010.09.033. Epub 2010 Sep 30. J Mol Biol. 2010. PMID: 20869366
-
Identification of residues required for stalled-ribosome rescue in the codon-independent release factor YaeJ.Nucleic Acids Res. 2014 Mar;42(5):3152-63. doi: 10.1093/nar/gkt1280. Epub 2013 Dec 9. Nucleic Acids Res. 2014. PMID: 24322300 Free PMC article.
-
Diversity and Similarity of Termination and Ribosome Rescue in Bacterial, Mitochondrial, and Cytoplasmic Translation.Biochemistry (Mosc). 2021 Sep;86(9):1107-1121. doi: 10.1134/S0006297921090066. Biochemistry (Mosc). 2021. PMID: 34565314 Free PMC article. Review.
-
The Origin and Evolution of Release Factors: Implications for Translation Termination, Ribosome Rescue, and Quality Control Pathways.Int J Mol Sci. 2019 Apr 23;20(8):1981. doi: 10.3390/ijms20081981. Int J Mol Sci. 2019. PMID: 31018531 Free PMC article. Review.
Cited by
-
Mitochondrial Protein Synthesis Adapts to Influx of Nuclear-Encoded Protein.Cell. 2016 Oct 6;167(2):471-483.e10. doi: 10.1016/j.cell.2016.09.003. Epub 2016 Sep 29. Cell. 2016. PMID: 27693358 Free PMC article.
-
The process of mammalian mitochondrial protein synthesis.Cell Tissue Res. 2017 Jan;367(1):5-20. doi: 10.1007/s00441-016-2456-0. Epub 2016 Jul 14. Cell Tissue Res. 2017. PMID: 27411691 Free PMC article. Review.
-
Evolution and diversification of the organellar release factor family.Mol Biol Evol. 2012 Nov;29(11):3497-512. doi: 10.1093/molbev/mss157. Epub 2012 Jun 11. Mol Biol Evol. 2012. PMID: 22688947 Free PMC article.
-
Leigh syndrome in a patient with a novel C12orf65 pathogenic variant: case report and literature review.Genet Mol Biol. 2020 May 29;43(2):e20180271. doi: 10.1590/1678-4685-GMB-2018-0271. eCollection 2020. Genet Mol Biol. 2020. PMID: 32478789 Free PMC article.
-
Resolving nonstop translation complexes is a matter of life or death.J Bacteriol. 2014 Jun;196(12):2123-30. doi: 10.1128/JB.01490-14. Epub 2014 Apr 4. J Bacteriol. 2014. PMID: 24706739 Free PMC article. Review.
References
-
- Chomyn A (1996) In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol 264: 197–211 - PubMed
-
- Chrzanowska-Lightowlers ZM, Preiss T, Lightowlers RN (1994) Inhibition of mitochondrial protein synthesis promotes increased stability of nuclear-encoded respiratory gene transcripts. J Biol Chem 269: 27322–27328 - PubMed
-
- Das G, Varshney U (2006) Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 152: 2191–2195 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

