Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction
- PMID: 20186995
- PMCID: PMC4340654
- DOI: 10.1002/jgm.1440
Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction
Abstract
Background: We recently expressed a potent and noncytotoxic short hairpin (sh)RNA directed against chemokine (c-c motif) receptor 5 (CCR5) using lentiviral mediated transduction of CD34+ hematopoietic progenitor cells (HPCs) and demonstrated the stable reduction of CCR5 expression in T-lymphocytes.
Methods: In the present study, we further assessed the activity of the shRNA through HPC transduction and differentiation into macrophages derived from fetal liver CD34+ (FL-CD34+) HPCs. Transduced lentiviral vector encoding the human CCR5 shRNA was stably maintained in FL-CD34+ cells and in the terminally differentiated macrophages using macrophage colony-stimulating factor, granulocyte macrophage colony-stimulating factor, interleukin-3 and stem cell factor.
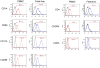
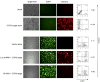
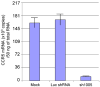
Results: Quantitative real-time polymerase chain reaction for CCR5 mRNA indicated over 90% reduction of CCR5 mRNA levels in CCR5 shRNA-transduced population. The cells with knockdown of CCR5 expression acquired resistance to R5 tropic HIV-1 NFN-SX strain. We also developed a novel approach utilizing a mCherry-CCR5 chimeric reporter to assess the effectiveness of CCR5 target down-regulation in macrophages directly. Both the shRNA and the reporter were maintained throughout HPC differentiation to macrophages without apparent cytotoxicity.
Conclusions: The present study demonstrates a novel method to simply and directly assess the function of small interfering RNA and the effective inhibition of HIV-1 infection by a potential potent shRNA to CCR5 delivered into macrophages derived from HPCs.
Figures
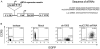




Similar articles
-
CXCR4 and CCR5 shRNA transgenic CD34+ cell derived macrophages are functionally normal and resist HIV-1 infection.Retrovirology. 2005 Aug 18;2:53. doi: 10.1186/1742-4690-2-53. Retrovirology. 2005. PMID: 16109172 Free PMC article.
-
Multivalent anti-CCR ribozymes for stem cell-based HIV type 1 gene therapy.AIDS Res Hum Retroviruses. 2001 Mar 20;17(5):385-99. doi: 10.1089/088922201750102427. AIDS Res Hum Retroviruses. 2001. PMID: 11282007
-
A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model.Blood. 2010 Feb 25;115(8):1534-44. doi: 10.1182/blood-2009-04-215855. Epub 2009 Dec 17. Blood. 2010. PMID: 20018916 Free PMC article.
-
siRNAs, ribozymes and RNA decoys in modeling stem cell-based gene therapy for HIV/AIDS.Anticancer Res. 2003 May-Jun;23(3A):1997-2005. Anticancer Res. 2003. PMID: 12894572 Review.
-
CCR5-targeted hematopoietic stem cell gene approaches for HIV disease: current progress and future prospects.Curr Stem Cell Res Ther. 2012 Jul;7(4):310-7. doi: 10.2174/157488812800793108. Curr Stem Cell Res Ther. 2012. PMID: 22486585 Review.
Cited by
-
CCR5 is a suppressor for cortical plasticity and hippocampal learning and memory.Elife. 2016 Dec 20;5:e20985. doi: 10.7554/eLife.20985. Elife. 2016. PMID: 27996938 Free PMC article.
-
Novel approaches to inhibit HIV entry.Viruses. 2012 Feb;4(2):309-24. doi: 10.3390/v4020309. Epub 2012 Feb 21. Viruses. 2012. PMID: 22470838 Free PMC article. Review.
-
Modeling Anti-HIV-1 HSPC-Based Gene Therapy in Humanized Mice Previously Infected with HIV-1.Mol Ther Methods Clin Dev. 2017 Dec 1;9:23-32. doi: 10.1016/j.omtm.2017.11.008. eCollection 2018 Jun 15. Mol Ther Methods Clin Dev. 2017. PMID: 29322065 Free PMC article.
-
Chemokine receptor 5 knockout strategies.Curr Opin HIV AIDS. 2011 Jan;6(1):74-9. doi: 10.1097/COH.0b013e32834122d7. Curr Opin HIV AIDS. 2011. PMID: 21242897 Free PMC article. Review.
-
Ectopic expression of anti-HIV-1 shRNAs protects CD8(+) T cells modified with CD4ζ CAR from HIV-1 infection and alleviates impairment of cell proliferation.Biochem Biophys Res Commun. 2015 Jul 31;463(3):216-21. doi: 10.1016/j.bbrc.2015.05.026. Epub 2015 May 19. Biochem Biophys Res Commun. 2015. PMID: 25998390 Free PMC article.
References
-
- Gulick RM, Mellors JW, Havlir D, et al. Simultaneous versus sequential initiation of therapy with indinavir, zidovudine, and lamivudine for HIV-1 infection: 100-week follow-up. JAMA. 1998;280:35–41. - PubMed
-
- Gulick RM, Mellors JW, Havlir D, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. New Engl J Med. 1997;337:734–739. - PubMed
-
- Hammer SM, Squires KE, Hughes MD, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. New Engl J Med. 1997;337:725–733. - PubMed
-
- Brau N, Leaf HL, Wieczorek RL, et al. Severe hepatitis in three AIDS patients treated with indinavir. Lancet. 1997;349:924–925. - PubMed
-
- d'Arminio Monforte A, Lepri AC, Rezza G, et al. Insights into the reasons for discontinuation of the first highly active antiretroviral therapy (HAART) regimen in a cohort of antiretroviral naive patients. ICONA Study Group. Italian Cohort of Antiretroviral-Naive Patients. AIDS. 2000;14:499–507. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

