A modular positive feedback-based gene amplifier
- PMID: 20187959
- PMCID: PMC2845093
- DOI: 10.1186/1754-1611-4-4
A modular positive feedback-based gene amplifier
Abstract
Background: Positive feedback is a common mechanism used in the regulation of many gene circuits as it can amplify the response to inducers and also generate binary outputs and hysteresis. In the context of electrical circuit design, positive feedback is often considered in the design of amplifiers. Similar approaches, therefore, may be used for the design of amplifiers in synthetic gene circuits with applications, for example, in cell-based sensors.
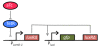
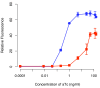
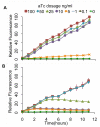
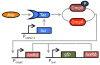
Results: We developed a modular positive feedback circuit that can function as a genetic signal amplifier, heightening the sensitivity to inducer signals as well as increasing maximum expression levels without the need for an external cofactor. The design utilizes a constitutively active, autoinducer-independent variant of the quorum-sensing regulator LuxR. We experimentally tested the ability of the positive feedback module to separately amplify the output of a one-component tetracycline sensor and a two-component aspartate sensor. In each case, the positive feedback module amplified the response to the respective inducers, both with regards to the dynamic range and sensitivity.
Conclusions: The advantage of our design is that the actual feedback mechanism depends only on a single gene and does not require any other modulation. Furthermore, this circuit can amplify any transcriptional signal, not just one encoded within the circuit or tuned by an external inducer. As our design is modular, it can potentially be used as a component in the design of more complex synthetic gene circuits.
Figures



Similar articles
-
Hysteretic Genetic Circuit for Detection of Proteasomal Degradation in Mammalian Cells.ACS Synth Biol. 2019 Sep 20;8(9):2025-2035. doi: 10.1021/acssynbio.9b00074. Epub 2019 Aug 27. ACS Synth Biol. 2019. PMID: 31415719
-
Robust and sensitive control of a quorum-sensing circuit by two interlocked feedback loops.Mol Syst Biol. 2008;4:234. doi: 10.1038/msb.2008.70. Epub 2008 Dec 16. Mol Syst Biol. 2008. PMID: 19096361 Free PMC article.
-
Design and mathematical analysis of activating transcriptional amplifiers that enable modular temporal control in synthetic juxtacrine circuits.Synth Syst Biotechnol. 2023 Oct 10;8(4):654-672. doi: 10.1016/j.synbio.2023.09.008. eCollection 2023 Dec. Synth Syst Biotechnol. 2023. PMID: 37868744 Free PMC article.
-
The Merging of Biological and Electronic Circuits.iScience. 2020 Oct 15;23(11):101688. doi: 10.1016/j.isci.2020.101688. eCollection 2020 Nov 20. iScience. 2020. PMID: 33163942 Free PMC article. Review.
-
Tools and Principles for Microbial Gene Circuit Engineering.J Mol Biol. 2016 Feb 27;428(5 Pt B):862-88. doi: 10.1016/j.jmb.2015.10.004. Epub 2015 Oct 20. J Mol Biol. 2016. PMID: 26463592 Review.
Cited by
-
Hidden hysteresis - population dynamics can obscure gene network dynamics.J Biol Eng. 2013 Jun 24;7(1):16. doi: 10.1186/1754-1611-7-16. J Biol Eng. 2013. PMID: 23800122 Free PMC article.
-
Tailor-made transcriptional biosensors for optimizing microbial cell factories.J Ind Microbiol Biotechnol. 2017 May;44(4-5):623-645. doi: 10.1007/s10295-016-1862-3. Epub 2016 Nov 11. J Ind Microbiol Biotechnol. 2017. PMID: 27837353 Review.
-
Characterization of an inducible promoter in different DNA copy number conditions.BMC Bioinformatics. 2012 Mar 28;13 Suppl 4(Suppl 4):S11. doi: 10.1186/1471-2105-13-S4-S11. BMC Bioinformatics. 2012. PMID: 22536957 Free PMC article.
-
Synthetic Biology Enables Programmable Cell-Based Biosensors.Chemphyschem. 2020 Jan 16;21(2):132-144. doi: 10.1002/cphc.201900739. Epub 2019 Oct 25. Chemphyschem. 2020. PMID: 31585026 Free PMC article. Review.
-
A positive feedback-based gene circuit to increase the production of a membrane protein.J Biol Eng. 2010 May 25;4:6. doi: 10.1186/1754-1611-4-6. J Biol Eng. 2010. PMID: 20500847 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

