Extended structures in RNA folding intermediates are due to nonnative interactions rather than electrostatic repulsion
- PMID: 20188108
- PMCID: PMC2873146
- DOI: 10.1016/j.jmb.2010.02.025
Extended structures in RNA folding intermediates are due to nonnative interactions rather than electrostatic repulsion
Abstract
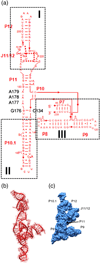
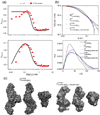
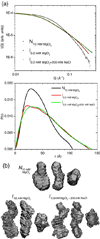
RNA folding occurs via a series of transitions between metastable intermediate states for Mg(2+) concentrations below those needed to fold the native structure. In general, these folding intermediates are considerably less compact than their respective native states. Our previous work demonstrates that the major equilibrium intermediate of the 154-residue specificity domain (S-domain) of the Bacillus subtilis RNase P RNA is more extended than its native structure. We now investigate two models with falsifiable predictions regarding the origins of the extended intermediate structures in the S-domains of the B. subtilis and the Escherichia coli RNase P RNA that belong to different classes of P RNA and have distinct native structures. The first model explores the contribution of electrostatic repulsion, while the second model probes specific interactions in the core of the folding intermediate. Using small-angle X-ray scattering and Langevin dynamics simulations, we show that electrostatics plays only a minor role, whereas specific interactions largely account for the extended nature of the intermediate. Structural contacts in the core, including a nonnative base pair, help to stabilize the intermediate conformation. We conclude that RNA folding intermediates adopt extended conformations due to short-range, nonnative interactions rather than generic electrostatic repulsion of helical domains. These principles apply to other ribozymes and riboswitches that undergo functionally relevant conformational changes.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures

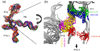
Similar articles
-
Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding.J Mol Biol. 2005 Sep 23;352(3):712-22. doi: 10.1016/j.jmb.2005.07.010. J Mol Biol. 2005. PMID: 16115647
-
Structural plasticity and Mg2+ binding properties of RNase P P4 from combined analysis of NMR residual dipolar couplings and motionally decoupled spin relaxation.RNA. 2007 Feb;13(2):251-66. doi: 10.1261/rna.264207. Epub 2006 Dec 28. RNA. 2007. PMID: 17194721 Free PMC article.
-
Mg2+-dependent compaction and folding of yeast tRNAPhe and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle X-ray scattering.Biochemistry. 2000 Sep 12;39(36):11107-13. doi: 10.1021/bi000724n. Biochemistry. 2000. PMID: 10998249
-
Structures of helical junctions in nucleic acids.Q Rev Biophys. 2000 May;33(2):109-59. doi: 10.1017/s0033583500003590. Q Rev Biophys. 2000. PMID: 11131562 Review.
-
RNA misfolding and the action of chaperones.Front Biosci. 2008 Jan 1;13:1-20. doi: 10.2741/2557. Front Biosci. 2008. PMID: 17981525 Free PMC article. Review.
Cited by
-
Footprinting analysis of interactions between the largest eukaryotic RNase P/MRP protein Pop1 and RNase P/MRP RNA components.RNA. 2015 Sep;21(9):1591-605. doi: 10.1261/rna.049007.114. Epub 2015 Jul 1. RNA. 2015. PMID: 26135751 Free PMC article.
-
Insights into the regulatory landscape of the lysine riboswitch.J Mol Biol. 2012 Oct 12;423(1):17-33. doi: 10.1016/j.jmb.2012.06.038. Epub 2012 Jul 3. J Mol Biol. 2012. PMID: 22771573 Free PMC article.
-
Many-body effect in ion binding to RNA.J Chem Phys. 2014 Aug 7;141(5):055101. doi: 10.1063/1.4890656. J Chem Phys. 2014. PMID: 25106614 Free PMC article.
-
Predicting translational diffusion of evolutionary conserved RNA structures by the nucleotide number.Nucleic Acids Res. 2011 Feb;39(3):e17. doi: 10.1093/nar/gkq808. Epub 2010 Nov 10. Nucleic Acids Res. 2011. PMID: 21068070 Free PMC article.
-
Folding of RNA tertiary structure: Linkages between backbone phosphates, ions, and water.Biopolymers. 2013 Dec;99(12):1105-13. doi: 10.1002/bip.22249. Biopolymers. 2013. PMID: 23568785 Free PMC article. Review.
References
-
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Ann. Rev. Biophys. Biomol. Struct. 1997;26:113–137. - PubMed
-
- Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. - PubMed
-
- Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. - PubMed
-
- Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. - PubMed
-
- Woodson SA. Folding mechanisms of group I ribozymes: role of stability and contact order. Biochem Soc Trans. 2002;30:1166–1169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

