Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader
- PMID: 20188667
- PMCID: PMC2830912
- DOI: 10.1016/j.molcel.2010.01.013
Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader
Abstract
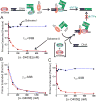
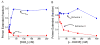
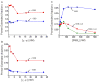
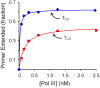
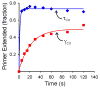
Cellular replicases contain multiprotein ATPases that load sliding clamp processivity factors onto DNA. We reveal an additional role for the DnaX clamp loader: chaperoning of the replicative polymerase onto a clamp newly bound to DNA. We show that chaperoning confers distinct advantages, including marked acceleration of initiation complex formation. We reveal a requirement for the tau form of DnaX complex to relieve inhibition by single-stranded DNA binding protein during initiation complex formation. We propose that, after loading beta(2), DnaX complex preserves an SSB-free segment of DNA immediately downstream of the primer terminus and chaperones Pol III into that position, preventing competition by SSB. The C-terminal tail of SSB stimulates reactions catalyzed by tau-containing DnaX complexes through a contact distinct from the contact involving the chi subunit. Chaperoning of Pol III by the DnaX complex provides a molecular explanation for how initiation complexes form when supported by the nonhydrolyzed analog ATPgammaS.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
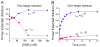

Similar articles
-
Polymerase chaperoning and multiple ATPase sites enable the E. coli DNA polymerase III holoenzyme to rapidly form initiation complexes.J Mol Biol. 2011 Sep 23;412(3):340-53. doi: 10.1016/j.jmb.2011.07.051. Epub 2011 Jul 28. J Mol Biol. 2011. PMID: 21820444 Free PMC article.
-
Only one ATP-binding DnaX subunit is required for initiation complex formation by the Escherichia coli DNA polymerase III holoenzyme.J Biol Chem. 2010 Sep 17;285(38):29049-53. doi: 10.1074/jbc.C110.165076. Epub 2010 Jul 30. J Biol Chem. 2010. PMID: 20675375 Free PMC article.
-
DNA Polymerase III, but Not Polymerase IV, Must Be Bound to a τ-Containing DnaX Complex to Enable Exchange into Replication Forks.J Biol Chem. 2016 May 27;291(22):11727-35. doi: 10.1074/jbc.M116.725358. Epub 2016 Apr 7. J Biol Chem. 2016. PMID: 27056333 Free PMC article.
-
Bacterial replicases and related polymerases.Curr Opin Chem Biol. 2011 Oct;15(5):587-94. doi: 10.1016/j.cbpa.2011.07.018. Epub 2011 Aug 19. Curr Opin Chem Biol. 2011. PMID: 21855395 Free PMC article. Review.
-
DNA replicases from a bacterial perspective.Annu Rev Biochem. 2011;80:403-36. doi: 10.1146/annurev-biochem-061208-091655. Annu Rev Biochem. 2011. PMID: 21675919 Review.
Cited by
-
Mutant DnaAs of Escherichia coli that are refractory to negative control.Nucleic Acids Res. 2013 Dec;41(22):10254-67. doi: 10.1093/nar/gkt774. Epub 2013 Aug 29. Nucleic Acids Res. 2013. PMID: 23990329 Free PMC article.
-
Slow unloading leads to DNA-bound β2-sliding clamp accumulation in live Escherichia coli cells.Nat Commun. 2014 Dec 18;5:5820. doi: 10.1038/ncomms6820. Nat Commun. 2014. PMID: 25520215 Free PMC article.
-
ssb gene duplication restores the viability of ΔholC and ΔholD Escherichia coli mutants.PLoS Genet. 2014 Oct 16;10(10):e1004719. doi: 10.1371/journal.pgen.1004719. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25329071 Free PMC article.
-
Insights into Okazaki fragment synthesis by the T4 replisome: the fate of lagging-strand holoenzyme components and their influence on Okazaki fragment size.J Biol Chem. 2013 Jul 19;288(29):20807-20816. doi: 10.1074/jbc.M113.485961. Epub 2013 May 31. J Biol Chem. 2013. PMID: 23729670 Free PMC article.
-
Stepwise assembly of the human replicative polymerase holoenzyme.Elife. 2013 Apr 2;2:e00278. doi: 10.7554/eLife.00278. Elife. 2013. PMID: 23577232 Free PMC article.
References
-
- Alley SC, Abel-Santos E, Benkovic SJ. Tracking Sliding Clamp Opening and Closing During Bacteriophage T4 DNA Polymerase Holoenzyme Assembly. Biochemistry. 2000;39:3076–3090. - PubMed
-
- Ason B, Bertram JG, Hingorani MM, Beechem JM, O'Donnell ME, Goodman MF, Bloom LB. A Model for Escherichia coli DNA Polymerase III Holoenzyme Assembly at Primer/Template Ends. DNA triggers a change in binding specificity of the γ complex clamp loader. J Biol Chem. 2000;275:3006–3015. - PubMed
-
- Bertram JG, Bloom LB, Turner J, O'Donnell ME, Beechem JM, Goodman MF. Pre-Steady State Analysis of the Assembly of Wild Type and Mutant Circular Clamps of Escherichia coli DNA Polymerase III onto DNA. J Biol Chem. 1998;273:24564–24574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

