Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair
- PMID: 20188668
- PMCID: PMC2958316
- DOI: 10.1016/j.molcel.2010.01.021
Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair
Abstract
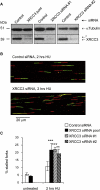
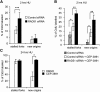
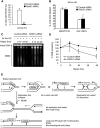
Faithful DNA replication is essential to all life. Hydroxyurea (HU) depletes the cells of dNTPs, which initially results in stalled replication forks that, after prolonged treatment, collapse into DSBs. Here, we report that stalled replication forks are efficiently restarted in a RAD51-dependent process that does not trigger homologous recombination (HR). The XRCC3 protein, which is required for RAD51 foci formation, is also required for replication restart of HU-stalled forks, suggesting that RAD51-mediated strand invasion supports fork restart. In contrast, replication forks collapsed by prolonged replication blocks do not restart, and global replication is rescued by new origin firing. We find that RAD51-dependent HR is triggered for repair of collapsed replication forks, without apparent restart. In conclusion, our data suggest that restart of stalled replication forks and HR repair of collapsed replication forks require two distinct RAD51-mediated pathways.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
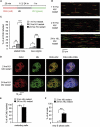
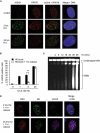
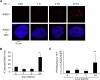

References
-
- Anglana M., Apiou F., Bensimon A., Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. - PubMed
-
- Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. - PubMed
-
- Baumann P., Benson F.E., West S.C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

