The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans
- PMID: 20188671
- PMCID: PMC2846537
- DOI: 10.1016/j.molcel.2010.01.015
The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans
Abstract
Genetic analyses previously implicated the matrix-localized protease ClpP in signaling the stress of protein misfolding in the mitochondrial matrix to activate nuclear-encoded mitochondrial chaperone genes in C. elegans (UPR(mt)). Here, we report that haf-1, a gene encoding a mitochondria-localized ATP-binding cassette protein, is required for signaling within the UPR(mt) and for coping with misfolded protein stress. Peptide efflux from isolated mitochondria was ATP dependent and required HAF-1 and the protease ClpP. Defective UPR(mt) signaling in the haf-1-deleted worms was associated with failure of the bZIP protein, ZC376.7, to localize to nuclei in worms with perturbed mitochondrial protein folding, whereas zc376.7(RNAi) strongly inhibited the UPR(mt). These observations suggest a simple model whereby perturbation of the protein-folding environment in the mitochondrial matrix promotes ClpP-mediated generation of peptides whose haf-1-dependent export from the matrix contributes to UPR(mt) signaling across the mitochondrial inner membrane.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


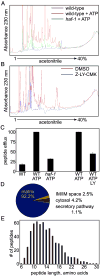


Comment in
-
Peptides signal mitochondrial stress.Cell Metab. 2010 Mar 3;11(3):177-8. doi: 10.1016/j.cmet.2010.02.011. Cell Metab. 2010. PMID: 20197049
References
-
- Arnold I, Wagner-Ecker M, Ansorge W, Langer T. Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene. 2006;367:74–88. - PubMed
-
- Augustin S, Nolden M, Muller S, Hardt O, Arnold I, Langer T. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem. 2005;280:2691–2699. - PubMed
-
- Barral JM, Broadley SA, Schaffar G, Hartl FU. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15:17–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

