Bacterial lifestyle in a deep-sea hydrothermal vent chimney revealed by the genome sequence of the thermophilic bacterium Deferribacter desulfuricans SSM1
- PMID: 20189949
- PMCID: PMC2885270
- DOI: 10.1093/dnares/dsq005
Bacterial lifestyle in a deep-sea hydrothermal vent chimney revealed by the genome sequence of the thermophilic bacterium Deferribacter desulfuricans SSM1
Abstract
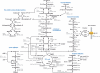
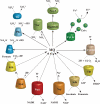
The complete genome sequence of the thermophilic sulphur-reducing bacterium, Deferribacter desulfuricans SMM1, isolated from a hydrothermal vent chimney has been determined. The genome comprises a single circular chromosome of 2,234,389 bp and a megaplasmid of 308,544 bp. Many genes encoded in the genome are most similar to the genes of sulphur- or sulphate-reducing bacterial species within Deltaproteobacteria. The reconstructed central metabolisms showed a heterotrophic lifestyle primarily driven by C1 to C3 organics, e.g. formate, acetate, and pyruvate, and also suggested that the inability of autotrophy via a reductive tricarboxylic acid cycle may be due to the lack of ATP-dependent citrate lyase. In addition, the genome encodes numerous genes for chemoreceptors, chemotaxis-like systems, and signal transduction machineries. These signalling networks may be linked to this bacterium's versatile energy metabolisms and may provide ecophysiological advantages for D. desulfuricans SSM1 thriving in the physically and chemically fluctuating environments near hydrothermal vents. This is the first genome sequence from the phylum Deferribacteres.
Figures



Similar articles
-
Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent.Int J Syst Evol Microbiol. 2003 May;53(Pt 3):839-846. doi: 10.1099/ijs.0.02479-0. Int J Syst Evol Microbiol. 2003. PMID: 12807210
-
Complete genome sequence of Crassaminicella sp. 143-21,isolated from a deep-sea hydrothermal vent.Mar Genomics. 2022 Apr;62:100899. doi: 10.1016/j.margen.2021.100899. Epub 2021 Sep 4. Mar Genomics. 2022. PMID: 35246304
-
Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate-ammonifying bacterium from deep-sea hydrothermal vents.Int J Syst Evol Microbiol. 2004 Jan;54(Pt 1):175-181. doi: 10.1099/ijs.0.02781-0. Int J Syst Evol Microbiol. 2004. PMID: 14742477
-
Complete genome sequence of Methanofervidicoccus sp. A16, a thermophilic methanogen isolated from Mid Cayman Rise hydrothermal vent.Mar Genomics. 2020 Oct;53:100768. doi: 10.1016/j.margen.2020.100768. Epub 2020 Mar 26. Mar Genomics. 2020. PMID: 32222383
-
An abyssal mobilome: viruses, plasmids and vesicles from deep-sea hydrothermal vents.Res Microbiol. 2015 Dec;166(10):742-52. doi: 10.1016/j.resmic.2015.04.001. Epub 2015 Apr 22. Res Microbiol. 2015. PMID: 25911507 Review.
Cited by
-
Genomic Insights into the Carbon and Energy Metabolism of a Thermophilic Deep-Sea Bacterium Deferribacter autotrophicus Revealed New Metabolic Traits in the Phylum Deferribacteres.Genes (Basel). 2019 Oct 26;10(11):849. doi: 10.3390/genes10110849. Genes (Basel). 2019. PMID: 31717820 Free PMC article.
-
Genome sequence of the moderately thermophilic halophile Flexistipes sinusarabici strain (MAS10).Stand Genomic Sci. 2011 Oct 15;5(1):86-96. doi: 10.4056/sigs.2235024. Epub 2011 Sep 23. Stand Genomic Sci. 2011. PMID: 22180813 Free PMC article.
-
Complete genome sequence of Denitrovibrio acetiphilus type strain (N2460).Stand Genomic Sci. 2010 Jun 15;2(3):270-9. doi: 10.4056/sigs.892105. Stand Genomic Sci. 2010. PMID: 21304711 Free PMC article.
-
Expanded Diversity and Phylogeny of mer Genes Broadens Mercury Resistance Paradigms and Reveals an Origin for MerA Among Thermophilic Archaea.Front Microbiol. 2021 Jun 23;12:682605. doi: 10.3389/fmicb.2021.682605. eCollection 2021. Front Microbiol. 2021. PMID: 34248899 Free PMC article.
-
Genomic evidence for the first symbiotic Deferribacterota, a novel gut symbiont from the deep-sea hydrothermal vent shrimp Rimicaris kairei.Front Microbiol. 2023 Jun 29;14:1179935. doi: 10.3389/fmicb.2023.1179935. eCollection 2023. Front Microbiol. 2023. PMID: 37455748 Free PMC article.
References
-
- Gold T. In: The Deep Hot Biosphere. Gold T., editor. New York: Copernicus Books; 2001. pp. 19–22.
-
- Takai K., Kobayashi H., Nealson K.H., Horikoshi K. Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2003;53:839–46. - PubMed
-
- Miroshnichenko M.L., Slobodkin A.I., Kostrikina N.A., et al. Deferribacter abyssi sp. nov., an anaerobic thermophile from deep-sea hydrothermal vents of the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 2003;53:1637–41. - PubMed
-
- Greene A.C., Patel B.K., Sheehy A.J. Deferribacter thermophilus gen. nov., sp. nov., a novel thermophilic manganese- and iron-reducing bacterium isolated from a petroleum reservoir. Int. J. Syst. Bacteriol. 1997;47:505–9. - PubMed
-
- Slobodkina G.B., Kolganova T.V., Chernyh N.A., Querellou J., Bonch-Osmolovskaya E.A., Slobodkin A.I. Deferribacter autotrophicus sp. nov., an iron(III)-reducing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2009;59:1508–12. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

