SOD1 mutations targeting surface hydrogen bonds promote amyotrophic lateral sclerosis without reducing apo-state stability
- PMID: 20189984
- PMCID: PMC2885233
- DOI: 10.1074/jbc.M109.086074
SOD1 mutations targeting surface hydrogen bonds promote amyotrophic lateral sclerosis without reducing apo-state stability
Abstract
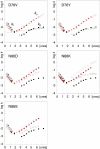
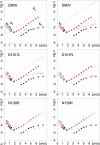
In good accord with the protein aggregation hypothesis for neurodegenerative diseases, ALS-associated SOD1 mutations are found to reduce structural stability or net repulsive charge. Moreover there are weak indications that the ALS disease progression rate is correlated with the degree of mutational impact on the apoSOD1 structure. A bottleneck for obtaining more conclusive information about these structure-disease relationships, however, is the large intrinsic variability in patient survival times and insufficient disease statistics for the majority of ALS-provoking mutations. As an alternative test of the structure-disease relationship we focus here on the SOD1 mutations that appear to be outliers in the data set. The results identify several ALS-provoking mutations whose only effect on apoSOD1 is the elimination or introduction of a single charge, i.e. D76V/Y, D101N, and N139D/K. The thermodynamic stability and folding behavior of these mutants are indistinguishable from the wild-type control. Moreover, D101N is an outlier in the plot of stability loss versus patient survival time by having rapid disease progression. Common to the identified mutations is that they truncate conserved salt-links and/or H-bond networks in the functional loops IV or VII. The results show that the local impact of ALS-associated mutations on the SOD1 molecule can sometimes overrun their global effects on apo-state stability and net repulsive charge, and point at the analysis of property outliers as an efficient strategy for mapping out new ALS-provoking features.
Figures

Similar articles
-
Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation.J Biol Chem. 2005 Apr 29;280(17):17266-74. doi: 10.1074/jbc.M500482200. Epub 2005 Feb 3. J Biol Chem. 2005. PMID: 15691826
-
Mechanical probes of SOD1 predict systematic trends in metal and dimer affinity of ALS-associated mutants.J Mol Biol. 2013 Mar 11;425(5):850-74. doi: 10.1016/j.jmb.2012.12.022. Epub 2013 Jan 3. J Mol Biol. 2013. PMID: 23291526
-
A double point mutation of SOD1 targeting net charge promotes aggregation under destabilizing conditions: Correlation of charge distribution and ALS-provoking mutation.Biochim Biophys Acta Gen Subj. 2023 May;1867(5):130325. doi: 10.1016/j.bbagen.2023.130325. Epub 2023 Feb 13. Biochim Biophys Acta Gen Subj. 2023. PMID: 36791828
-
SOD1 aggregation and ALS: role of metallation states and disulfide status.Curr Top Med Chem. 2012;12(22):2560-72. doi: 10.2174/1568026611212220010. Curr Top Med Chem. 2012. PMID: 23339308 Review.
-
Protein misfolding in the late-onset neurodegenerative diseases: common themes and the unique case of amyotrophic lateral sclerosis.Proteins. 2013 Aug;81(8):1285-303. doi: 10.1002/prot.24285. Epub 2013 Jul 2. Proteins. 2013. PMID: 23508986 Review.
Cited by
-
Decreased stability and increased formation of soluble aggregates by immature superoxide dismutase do not account for disease severity in ALS.Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2210-5. doi: 10.1073/pnas.0913021108. Epub 2011 Jan 21. Proc Natl Acad Sci U S A. 2011. PMID: 21257910 Free PMC article.
-
Early steps in thermal unfolding of superoxide dismutase 1 are similar to the conformational changes associated with the ALS-associated A4V mutation.Protein Eng Des Sel. 2013 Aug;26(8):503-13. doi: 10.1093/protein/gzt030. Epub 2013 Jun 19. Protein Eng Des Sel. 2013. PMID: 23784844 Free PMC article.
-
Genotype-property patient-phenotype relations suggest that proteome exhaustion can cause amyotrophic lateral sclerosis.PLoS One. 2015 Mar 23;10(3):e0118649. doi: 10.1371/journal.pone.0118649. eCollection 2015. PLoS One. 2015. PMID: 25798606 Free PMC article.
-
Species-specific activation of Cu/Zn SOD by its CCS copper chaperone in the pathogenic yeast Candida albicans.J Biol Inorg Chem. 2014 Jun;19(4-5):595-603. doi: 10.1007/s00775-013-1045-x. Epub 2013 Sep 17. J Biol Inorg Chem. 2014. PMID: 24043471 Free PMC article.
-
Nucleation and kinetics of SOD1 aggregation in human cells for ALS1.Mol Cell Biochem. 2020 Mar;466(1-2):117-128. doi: 10.1007/s11010-020-03693-y. Epub 2020 Feb 13. Mol Cell Biochem. 2020. PMID: 32056106
References
-
- Chiti F., Stefani M., Taddei N., Ramponi G., Dobson C. M. (2003) Nature 424, 805–808 - PubMed
-
- Alsod (2009) The ALS Online Database, http://alsod.iop.kcl.ac.uk/Als/index.aspx
-
- Sandelin E., Nordlund A., Andersen P. M., Marklund S. S., Oliveberg M. (2007) J. Biol. Chem. 282, 21230–21236 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

