Identification of a dehydrogenase required for lactose metabolism in Caulobacter crescentus
- PMID: 20190087
- PMCID: PMC2863468
- DOI: 10.1128/AEM.02085-09
Identification of a dehydrogenase required for lactose metabolism in Caulobacter crescentus
Abstract
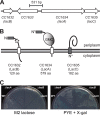
Caulobacter crescentus, which thrives in freshwater environments with low nutrient levels, serves as a model system for studying bacterial cell cycle regulation and organelle development. We examined its ability to utilize lactose (i) to gain insight into the metabolic capacities of oligotrophic bacteria and (ii) to obtain an additional genetic tool for studying this model organism, aiming to eliminate the basal enzymatic activity that hydrolyzes the chromogenic substrate 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-gal). Using a previously isolated transposon mutant, we identified a gene, lacA, that is required for growth on lactose as the sole carbon source and for turning colonies blue in the presence of X-gal. LacA, which contains a glucose-methanol-choline (GMC) oxidoreductase domain, has homology to the flavin subunit of Pectobacterium cypripedii's gluconate dehydrogenase. Sequence comparisons indicated that two genes near lacA, lacB and lacC, encode the other subunits of the membrane-bound dehydrogenase. In addition to lactose, all three lac genes are involved in the catabolism of three other beta-galactosides (lactulose, lactitol, and methyl-beta-d-galactoside) and two glucosides (salicin and trehalose). Dehydrogenase assays confirmed that the lac gene products oxidize lactose, salicin, and trehalose. This enzymatic activity is inducible, and increased lac expression in the presence of lactose and salicin likely contributes to the induction. Expression of lacA also depends on the presence of the lac genes, implying that the dehydrogenase participates in induction. The involvement of a dehydrogenase suggests that degradation of lactose and other sugars in C. crescentus may resemble a proposed pathway in Agrobacterium tumefaciens.
Figures
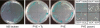
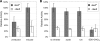


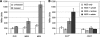
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1998. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
-
- Baldrian, P., and V. Valaskova. 2008. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 32:501-521. - PubMed
-
- Bastedo, D. P., and G. T. Marczynski. 2009. CtrA response regulator binding to the Caulobacter chromosome replication origin is required during nutrient and antibiotic stress as well as during cell cycle progression. Mol. Microbiol. 72:139-154. - PubMed
-
- Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

