A proteome-wide analysis of kinase-substrate network in the DNA damage response
- PMID: 20190278
- PMCID: PMC2857137
- DOI: 10.1074/jbc.M110.106989
A proteome-wide analysis of kinase-substrate network in the DNA damage response
Abstract
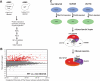
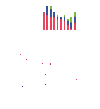
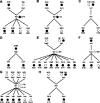
The DNA damage checkpoint, consisting of an evolutionarily conserved protein kinase cascade, controls the DNA damage response in eukaryotes. Knowledge of the in vivo substrates of the checkpoint kinases is essential toward understanding their functions. Here we used quantitative mass spectrometry to identify 53 new and 34 previously known targets of Mec1/Tel1, Rad53, and Dun1 in Saccharomyces cerevisiae. Analysis of replication protein A (RPA)-associated proteins reveals extensive physical interactions between RPA-associated proteins and Mec1/Tel1-specific substrates. Among them, multiple subunits of the chromatin remodeling complexes including ISW1, ISW2, INO80, SWR1, RSC, and SWI/SNF are identified and they undergo DNA damage-induced phosphorylation by Mec1 and Tel1. Taken together, this study greatly expands the existing knowledge of the targets of DNA damage checkpoint kinases and provides insights into the role of RPA-associated chromatins in mediating Mec1 and Tel1 substrate phosphorylation in vivo.
Figures

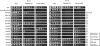
References
-
- Harrison J. C., Haber J. E. (2006) Annu. Rev. Genet 40, 209–235 - PubMed
-
- Kolodner R. D., Putnam C. D., Myung K. (2002) Science 297, 552–557 - PubMed
-
- Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. (2002) Annu. Rev. Genet 36, 617–656 - PubMed
-
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 - PubMed
-
- Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell Proteomics 1, 376–386 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

