Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia
- PMID: 20190740
- PMCID: PMC2989719
- DOI: 10.1038/ncb2037
Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia
Abstract
MicroRNAs (miRNAs) have emerged as novel cancer genes. In particular, the miR-17-92 cluster, containing six individual miRNAs, is highly expressed in haematopoietic cancers and promotes lymphomagenesis in vivo. Clinical use of these findings hinges on isolating the oncogenic activity within the 17-92 cluster and defining its relevant target genes. Here we show that miR-19 is sufficient to promote leukaemogenesis in Notch1-induced T-cell acute lymphoblastic leukaemia (T-ALL) in vivo. In concord with the pathogenic importance of this interaction in T-ALL, we report a novel translocation that targets the 17-92 cluster and coincides with a second rearrangement that activates Notch1. To identify the miR-19 targets responsible for its oncogenic action, we conducted a large-scale short hairpin RNA screen for genes whose knockdown can phenocopy miR-19. Strikingly, the results of this screen were enriched for miR-19 target genes, and include Bim (Bcl2L11), AMP-activated kinase (Prkaa1) and the phosphatases Pten and PP2A (Ppp2r5e). Hence, an unbiased, functional genomics approach reveals a coordinate clampdown on several regulators of phosphatidylinositol-3-OH kinase-related survival signals by the leukaemogenic miR-19.
Conflict of interest statement
Figures
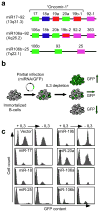
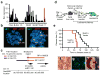
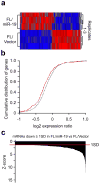
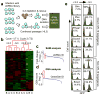
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

