Atomic accuracy in predicting and designing noncanonical RNA structure
- PMID: 20190761
- PMCID: PMC2854559
- DOI: 10.1038/nmeth.1433
Atomic accuracy in predicting and designing noncanonical RNA structure
Abstract
We present fragment assembly of RNA with full-atom refinement (FARFAR), a Rosetta framework for predicting and designing noncanonical motifs that define RNA tertiary structure. In a test set of thirty-two 6-20-nucleotide motifs, FARFAR recapitulated 50% of the experimental structures at near-atomic accuracy. Sequence redesign calculations recovered native bases at 65% of residues engaged in noncanonical interactions, and we experimentally validated mutations predicted to stabilize a signal recognition particle domain.
Figures
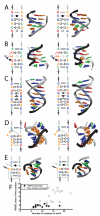
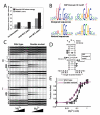
Comment in
-
Toward atomic accuracy in RNA design.Nat Methods. 2010 Apr;7(4):272-3. doi: 10.1038/nmeth0410-272. Nat Methods. 2010. PMID: 20354517
Similar articles
-
Modeling Small Noncanonical RNA Motifs with the Rosetta FARFAR Server.Methods Mol Biol. 2016;1490:187-98. doi: 10.1007/978-1-4939-6433-8_12. Methods Mol Biol. 2016. PMID: 27665600 Free PMC article.
-
Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.J Mol Biol. 2002 Mar 15;317(1):145-58. doi: 10.1006/jmbi.2002.5411. J Mol Biol. 2002. PMID: 11916385
-
Toward atomic accuracy in RNA design.Nat Methods. 2010 Apr;7(4):272-3. doi: 10.1038/nmeth0410-272. Nat Methods. 2010. PMID: 20354517
-
RNA structural motifs: building blocks of a modular biomolecule.Q Rev Biophys. 2005 Aug;38(3):221-43. doi: 10.1017/S0033583506004215. Epub 2006 Jul 3. Q Rev Biophys. 2005. PMID: 16817983 Review.
-
RNA structure prediction: an overview of methods.Methods Mol Biol. 2012;905:99-122. doi: 10.1007/978-1-61779-949-5_8. Methods Mol Biol. 2012. PMID: 22736001 Review.
Cited by
-
Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE).PLoS One. 2013 May 22;8(5):e63906. doi: 10.1371/journal.pone.0063906. Print 2013. PLoS One. 2013. PMID: 23717507 Free PMC article.
-
3dRNA: Building RNA 3D structure with improved template library.Comput Struct Biotechnol J. 2020 Aug 28;18:2416-2423. doi: 10.1016/j.csbj.2020.08.017. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33005304 Free PMC article.
-
Blind tests of RNA nearest-neighbor energy prediction.Proc Natl Acad Sci U S A. 2016 Jul 26;113(30):8430-5. doi: 10.1073/pnas.1523335113. Epub 2016 Jul 8. Proc Natl Acad Sci U S A. 2016. PMID: 27402765 Free PMC article.
-
Length-Dependent Deep Learning Model for RNA Secondary Structure Prediction.Molecules. 2022 Feb 2;27(3):1030. doi: 10.3390/molecules27031030. Molecules. 2022. PMID: 35164295 Free PMC article.
-
Classification and clustering of RNA crosslink-ligation data reveal complex structures and homodimers.Genome Res. 2022 May;32(5):968-985. doi: 10.1101/gr.275979.121. Epub 2022 Mar 24. Genome Res. 2022. PMID: 35332099 Free PMC article.
References
-
- Gesteland RF, Cech TR, Atkins JF. The RNA world : the nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006.
-
- Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Curr Opin Struct Biol. 2007 - PubMed
-
- Moore PB. Annu Rev Biochem. 1999;68:287. - PubMed
-
- Brion P, Westhof E. Annu Rev Biophys Biomol Struct. 1997;26:113. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

