A chemical and phosphoproteomic characterization of dasatinib action in lung cancer
- PMID: 20190765
- PMCID: PMC2842457
- DOI: 10.1038/nchembio.332
A chemical and phosphoproteomic characterization of dasatinib action in lung cancer
Abstract
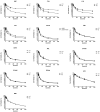
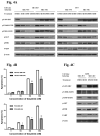
We describe a strategy for comprehending signaling pathways that are active in lung cancer cells and that are targeted by dasatinib using chemical proteomics to identify direct interacting proteins combined with immunoaffinity purification of tyrosine-phosphorylated peptides corresponding to activated tyrosine kinases. We identified nearly 40 different kinase targets of dasatinib. These include SRC-family kinase (SFK) members (LYN, SRC, FYN, LCK and YES), nonreceptor tyrosine kinases (FRK, BRK and ACK) and receptor tyrosine kinases (Ephrin receptors, DDR1 and EGFR). Using quantitative phosphoproteomics, we identified peptides corresponding to autophosphorylation sites of these tyrosine kinases that are inhibited in a concentration-dependent manner by dasatinib. Using drug-resistant gatekeeper mutants, we show that SFKs (particularly SRC and FYN), as well as EGFR, are relevant targets for dasatinib action. The combined mass spectrometry-based approach described here provides a system-level view of dasatinib action in cancer cells and suggests both functional targets and a rationale for combinatorial therapeutic strategies.
Figures



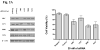
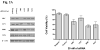

References
-
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–65. - PubMed
-
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. - PubMed
-
- Puri N, Khramtsov A, Ahmed S, et al. A selective small molecule inhibitor of c-Met, PHA665752, inhibits tumorigenicity and angiogenesis in mouse lung cancer xenografts. Cancer Res. 2007;67:3529–34. - PubMed
-
- Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006;66:5542–8. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

