Hidden Markov Models and their Applications in Biological Sequence Analysis
- PMID: 20190955
- PMCID: PMC2766791
- DOI: 10.2174/138920209789177575
Hidden Markov Models and their Applications in Biological Sequence Analysis
Abstract
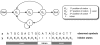
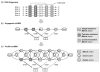
Hidden Markov models (HMMs) have been extensively used in biological sequence analysis. In this paper, we give a tutorial review of HMMs and their applications in a variety of problems in molecular biology. We especially focus on three types of HMMs: the profile-HMMs, pair-HMMs, and context-sensitive HMMs. We show how these HMMs can be used to solve various sequence analysis problems, such as pairwise and multiple sequence alignments, gene annotation, classification, similarity search, and many others.
Keywords: Hidden Markov model (HMM); context-sensitive HMM (csHMM); pair-HMM; profile-HMM; profile-csHMM; sequence analysis..
Figures



Similar articles
-
Applications of generalized pair hidden Markov models to alignment and gene finding problems.J Comput Biol. 2002;9(2):389-99. doi: 10.1089/10665270252935520. J Comput Biol. 2002. PMID: 12015888
-
Enhancing the quality of phylogenetic analysis using fuzzy hidden Markov model alignments.Stud Health Technol Inform. 2007;129(Pt 2):1245-9. Stud Health Technol Inform. 2007. PMID: 17911914
-
Fuzzy Hidden Markov Models: a new approach in multiple sequence alignment.Stud Health Technol Inform. 2006;124:99-104. Stud Health Technol Inform. 2006. PMID: 17108510
-
Profile hidden Markov models.Bioinformatics. 1998;14(9):755-63. doi: 10.1093/bioinformatics/14.9.755. Bioinformatics. 1998. PMID: 9918945 Review.
-
Rational Design of Profile Hidden Markov Models for Viral Classification and Discovery.In: Helder I. N, editor. Bioinformatics [Internet]. Brisbane (AU): Exon Publications; 2021 Mar 20. Chapter 9. In: Helder I. N, editor. Bioinformatics [Internet]. Brisbane (AU): Exon Publications; 2021 Mar 20. Chapter 9. PMID: 33877768 Free Books & Documents. Review.
Cited by
-
LAceP: lysine acetylation site prediction using logistic regression classifiers.PLoS One. 2014 Feb 20;9(2):e89575. doi: 10.1371/journal.pone.0089575. eCollection 2014. PLoS One. 2014. PMID: 24586884 Free PMC article.
-
Machine Learning and Deep Learning in Synthetic Biology: Key Architectures, Applications, and Challenges.ACS Omega. 2024 Feb 19;9(9):9921-9945. doi: 10.1021/acsomega.3c05913. eCollection 2024 Mar 5. ACS Omega. 2024. PMID: 38463314 Free PMC article. Review.
-
An Introduction to Infinite HMMs for Single-Molecule Data Analysis.Biophys J. 2017 May 23;112(10):2021-2029. doi: 10.1016/j.bpj.2017.04.027. Biophys J. 2017. PMID: 28538142 Free PMC article. Review.
-
Metagenomic Classification Using an Abstraction Augmented Markov Model.J Comput Biol. 2016 Feb;23(2):111-122. doi: 10.1089/cmb.2015.0141. Epub 2015 Nov 30. J Comput Biol. 2016. PMID: 26618474 Free PMC article.
-
HiMMe: using genetic patterns as a proxy for genome assembly reliability assessment.BMC Genomics. 2017 Sep 5;18(1):694. doi: 10.1186/s12864-017-3965-2. BMC Genomics. 2017. PMID: 28874136 Free PMC article.
References
-
- Rabiner L R. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286.
-
- Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis. Cambridge, UK: Cambridge University Press; 1998.
-
- Pachter L, Alexandersson M, Cawley S. Applications of generalized pair hidden Markov models to alignment and gene finding problems. J. Comput. Biol. 2002;9:389–399. - PubMed
-
- Liang K C, Wang X, Anastassiou D. Bayesian basecalling for DNA sequence analysis using hidden Markov models. IEEE/ACM Trans. Comput. Biol. Bioinform. 2007;4:430–440. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
