Compact intermediates in RNA folding
- PMID: 20192764
- PMCID: PMC6341483
- DOI: 10.1146/annurev.biophys.093008.131334
Compact intermediates in RNA folding
Abstract
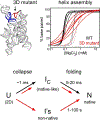
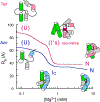
Large noncoding RNAs fold into their biologically functional structures via compact yet disordered intermediates, which couple the stable secondary structure of the RNA with the emerging tertiary fold. The specificity of the collapse transition, which coincides with the assembly of helical domains, depends on RNA sequence and counterions. It determines the specificity of the folding pathways and the magnitude of the free energy barriers to the ensuing search for the native conformation. By coupling helix assembly with nascent tertiary interactions, compact folding intermediates in RNA also play a crucial role in ligand binding and RNA-protein recognition.
Figures
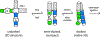
Similar articles
-
Cooperative tertiary interaction network guides RNA folding.Cell. 2012 Apr 13;149(2):348-57. doi: 10.1016/j.cell.2012.01.057. Cell. 2012. PMID: 22500801 Free PMC article.
-
Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering.J Am Chem Soc. 2010 Jul 28;132(29):10148-54. doi: 10.1021/ja103867p. J Am Chem Soc. 2010. PMID: 20597502 Free PMC article.
-
Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme.Proc Natl Acad Sci U S A. 2003 Feb 18;100(4):1574-9. doi: 10.1073/pnas.0337743100. Epub 2003 Feb 6. Proc Natl Acad Sci U S A. 2003. PMID: 12574513 Free PMC article.
-
Maximizing RNA folding rates: a balancing act.RNA. 2000 Jun;6(6):790-4. doi: 10.1017/s1355838200000522. RNA. 2000. PMID: 10864039 Free PMC article. Review.
-
Mapping nucleic acid structure by hydroxyl radical cleavage.Curr Opin Chem Biol. 2005 Apr;9(2):127-34. doi: 10.1016/j.cbpa.2005.02.009. Curr Opin Chem Biol. 2005. PMID: 15811796 Review.
Cited by
-
Spontaneous binding of single-stranded RNAs to RRM proteins visualized by unbiased atomistic simulations with a rescaled RNA force field.Nucleic Acids Res. 2022 Nov 28;50(21):12480-12496. doi: 10.1093/nar/gkac1106. Nucleic Acids Res. 2022. PMID: 36454011 Free PMC article.
-
The right angle (RA) motif: a prevalent ribosomal RNA structural pattern found in group I introns.J Mol Biol. 2012 Nov 23;424(1-2):54-67. doi: 10.1016/j.jmb.2012.09.012. Epub 2012 Sep 18. J Mol Biol. 2012. PMID: 22999957 Free PMC article.
-
Computational analysis of RNA structures with chemical probing data.Methods. 2015 Jun;79-80:60-6. doi: 10.1016/j.ymeth.2015.02.003. Epub 2015 Feb 14. Methods. 2015. PMID: 25687190 Free PMC article. Review.
-
The expression platform and the aptamer: cooperativity between Mg2+ and ligand in the SAM-I riboswitch.Nucleic Acids Res. 2013 Feb 1;41(3):1922-35. doi: 10.1093/nar/gks978. Epub 2012 Dec 20. Nucleic Acids Res. 2013. PMID: 23258703 Free PMC article.
-
Ion-mediated RNA structural collapse: effect of spatial confinement.Biophys J. 2012 Aug 22;103(4):827-36. doi: 10.1016/j.bpj.2012.06.048. Biophys J. 2012. PMID: 22947944 Free PMC article.
References
-
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

