Global dynamics of proteins: bridging between structure and function
- PMID: 20192781
- PMCID: PMC2938190
- DOI: 10.1146/annurev.biophys.093008.131258
Global dynamics of proteins: bridging between structure and function
Abstract
Biomolecular systems possess unique, structure-encoded dynamic properties that underlie their biological functions. Recent studies indicate that these dynamic properties are determined to a large extent by the topology of native contacts. In recent years, elastic network models used in conjunction with normal mode analyses have proven to be useful for elucidating the collective dynamics intrinsically accessible under native state conditions, including in particular the global modes of motions that are robustly defined by the overall architecture. With increasing availability of structural data for well-studied proteins in different forms (liganded, complexed, or free), there is increasing evidence in support of the correspondence between functional changes in structures observed in experiments and the global motions predicted by these coarse-grained analyses. These observed correlations suggest that computational methods may be advantageously employed for assessing functional changes in structure and allosteric mechanisms intrinsically favored by the native fold.
Figures
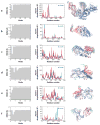
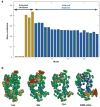
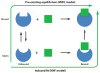

Similar articles
-
Structure-Encoded Global Motions and Their Role in Mediating Protein-Substrate Interactions.Biophys J. 2015 Sep 15;109(6):1101-9. doi: 10.1016/j.bpj.2015.06.004. Epub 2015 Jul 2. Biophys J. 2015. PMID: 26143655 Free PMC article. Review.
-
Pre-existing soft modes of motion uniquely defined by native contact topology facilitate ligand binding to proteins.Protein Sci. 2011 Oct;20(10):1645-58. doi: 10.1002/pro.711. Epub 2011 Sep 9. Protein Sci. 2011. PMID: 21826755 Free PMC article. Review.
-
Large-scale comparison of protein essential dynamics from molecular dynamics simulations and coarse-grained normal mode analyses.Proteins. 2010 Dec;78(16):3341-52. doi: 10.1002/prot.22841. Proteins. 2010. PMID: 20848551
-
Protein promiscuity: drug resistance and native functions--HIV-1 case.J Biomol Struct Dyn. 2005 Jun;22(6):615-24. doi: 10.1080/07391102.2005.10531228. J Biomol Struct Dyn. 2005. PMID: 15842167
-
Coevolution of function and the folding landscape: correlation with density of native contacts.Biophys J. 2008 Nov 1;95(9):L57-9. doi: 10.1529/biophysj.108.143388. Epub 2008 Aug 15. Biophys J. 2008. PMID: 18708465 Free PMC article.
Cited by
-
Characterization of the flexible lip regions in bacteriophage lambda lysozyme using MD simulations.Eur Biophys J. 2015 May;44(4):235-47. doi: 10.1007/s00249-015-1018-9. Epub 2015 Mar 28. Eur Biophys J. 2015. PMID: 25820531
-
Calculation of centralities in protein kinase A.Proc Natl Acad Sci U S A. 2022 Nov 22;119(47):e2215420119. doi: 10.1073/pnas.2215420119. Epub 2022 Nov 14. Proc Natl Acad Sci U S A. 2022. PMID: 36375071 Free PMC article.
-
Identification of motions in membrane proteins by elastic network models and their experimental validation.Methods Mol Biol. 2012;914:285-317. doi: 10.1007/978-1-62703-023-6_17. Methods Mol Biol. 2012. PMID: 22976035 Free PMC article.
-
Tunable microsecond dynamics of an allosteric switch regulate the activity of a AAA+ disaggregation machine.Nat Commun. 2019 Mar 29;10(1):1438. doi: 10.1038/s41467-019-09474-6. Nat Commun. 2019. PMID: 30926805 Free PMC article.
-
Large collective motions regulate the functional properties of glutamate transporter trimers.Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):15141-6. doi: 10.1073/pnas.1112216108. Epub 2011 Aug 29. Proc Natl Acad Sci U S A. 2011. PMID: 21876140 Free PMC article.
References
-
- Bahar I, Atilgan AR, Demirel MC, Erman B. Vibrational dynamics of folded proteins: significance of slow and fast motions in relation to function and stability. Phys Rev Lett. 1998;80:2733–36.
-
- Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

