Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases
- PMID: 20194115
- PMCID: PMC2896518
- DOI: 10.1093/nar/gkq122
Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases
Abstract
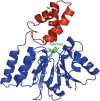
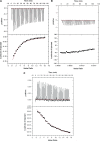
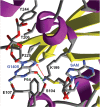
Sgm (Sisomicin-gentamicin methyltransferase) from antibiotic-producing bacterium Micromonospora zionensis is an enzyme that confers resistance to aminoglycosides like gentamicin and sisomicin by specifically methylating G1405 in bacterial 16S rRNA. Sgm belongs to the aminoglycoside resistance methyltransferase (Arm) family of enzymes that have been recently found to spread by horizontal gene transfer among disease-causing bacteria. Structural characterization of Arm enzymes is the key to understand their mechanism of action and to develop inhibitors that would block their activity. Here we report the structure of Sgm in complex with cofactors S-adenosylmethionine (AdoMet) and S-adenosylhomocysteine (AdoHcy) at 2.0 and 2.1 A resolution, respectively, and results of mutagenesis and rRNA footprinting, and protein-substrate docking. We propose the mechanism of methylation of G1405 by Sgm and compare it with other m(7)G methyltransferases, revealing a surprising diversity of active sites and binding modes for the same basic reaction of RNA modification. This analysis can serve as a stepping stone towards developing drugs that would specifically block the activity of Arm methyltransferases and thereby re-sensitize pathogenic bacteria to aminoglycoside antibiotics.
Figures

Similar articles
-
Functionally critical residues in the aminoglycoside resistance-associated methyltransferase RmtC play distinct roles in 30S substrate recognition.J Biol Chem. 2019 Nov 15;294(46):17642-17653. doi: 10.1074/jbc.RA119.011181. Epub 2019 Oct 8. J Biol Chem. 2019. PMID: 31594862 Free PMC article.
-
Critical residues for cofactor binding and catalytic activity in the aminoglycoside resistance methyltransferase Sgm.J Bacteriol. 2008 Sep;190(17):5855-61. doi: 10.1128/JB.00076-08. Epub 2008 Jun 27. J Bacteriol. 2008. PMID: 18586937 Free PMC article.
-
Modeling and experimental analyses reveal a two-domain structure and amino acids important for the activity of aminoglycoside resistance methyltransferase Sgm.Biochim Biophys Acta. 2008 Apr;1784(4):582-90. doi: 10.1016/j.bbapap.2007.09.009. Epub 2007 Sep 29. Biochim Biophys Acta. 2008. PMID: 18343347
-
Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases.Infect Dis Clin North Am. 2016 Jun;30(2):523-537. doi: 10.1016/j.idc.2016.02.011. Infect Dis Clin North Am. 2016. PMID: 27208771 Free PMC article. Review.
-
Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update.Drug Resist Updat. 2012 Jun;15(3):133-48. doi: 10.1016/j.drup.2012.05.001. Epub 2012 Jun 4. Drug Resist Updat. 2012. PMID: 22673098 Review.
Cited by
-
Substrate Recognition and Modification by a Pathogen-Associated Aminoglycoside Resistance 16S rRNA Methyltransferase.Antimicrob Agents Chemother. 2017 Apr 24;61(5):e00077-17. doi: 10.1128/AAC.00077-17. Print 2017 May. Antimicrob Agents Chemother. 2017. PMID: 28289026 Free PMC article.
-
Biological roles of RNA m7G modification and its implications in cancer.Biol Direct. 2023 Sep 14;18(1):58. doi: 10.1186/s13062-023-00414-5. Biol Direct. 2023. PMID: 37710294 Free PMC article. Review.
-
The Pathogen-Derived Aminoglycoside Resistance 16S rRNA Methyltransferase NpmA Possesses Dual m1A1408/m1G1408 Specificity.Antimicrob Agents Chemother. 2015 Dec;59(12):7862-5. doi: 10.1128/AAC.01872-15. Epub 2015 Sep 28. Antimicrob Agents Chemother. 2015. PMID: 26416864 Free PMC article.
-
Expression, purification and crystallization of adenosine 1408 aminoglycoside-resistance rRNA methyltransferases for structural studies.Protein Expr Purif. 2011 Jan;75(1):89-94. doi: 10.1016/j.pep.2010.07.005. Epub 2010 Jul 25. Protein Expr Purif. 2011. PMID: 20667473 Free PMC article.
-
Properties of small rRNA methyltransferase RsmD: mutational and kinetic study.RNA. 2012 Jun;18(6):1178-85. doi: 10.1261/rna.032763.112. Epub 2012 Apr 25. RNA. 2012. PMID: 22535590 Free PMC article.

