Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction
- PMID: 20194324
- PMCID: PMC2827826
- DOI: 10.2106/JBJS.I.00400
Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction
Abstract
Background: Previously described molecular biology techniques used to detect periprosthetic infections have been complicated by false-positive results. We have reported the development of a messenger RNA (mRNA)-based procedure to reduce these false-positive results. The limitations of this procedure are the lack of a universal target and reduced sensitivity due to a low concentration of bacterial mRNAs in test samples. The objective of the present study was to determine whether reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using universal primers can be used to detect the more abundant bacterial ribosomal RNA (rRNA) as an indicator of periprosthetic infection.
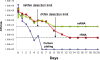
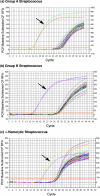
Methods: Serial dilutions of simulated synovial fluid infections were analyzed with rRNA RT-qPCR to determine the detection limit of this assay. Escherichia coli cultures treated with gentamicin were analyzed with RT-qPCR over a twenty-day time course to determine the degradation of rRNA as compared with the decrease in the viable cell count as determined by means of cell plating. As a proof of concept, group-specific polymerase chain reaction primers were developed for Streptococcus species and were tested against fifteen orthopaedically relevant organisms to show the potential for speciation with this assay. Sixty-four patients with a symptomatic effusion at the site of a total knee arthroplasty were enrolled, and complete patient information was documented in a prospective manner. Synovial fluid analysis with rRNA RT-qPCR was performed in a blind fashion.
Results: The rRNA RT-qPCR assay was able to detect as few as 590 colony forming units/mL of Staphylococcus aureus and 2900 colony forming units/mL of Escherichia coli. The rRNA RT-qPCR signal closely followed cell death, pointing to its potential use as a viability marker. Three group-specific primer sets correctly identified their intended targets without amplifying closely related species. Clinically, the test correctly identified all six patients with a confirmed infection and all fifty patients who clearly did not have an infection. Eight patients had some laboratory or clinical signs of infection, but their status could not be confirmed. Infection was indicated by rRNA RT-qPCR in three of these patients who had elevated synovial fluid white blood-cell counts but negative results on culture. For statistical purposes, all patients who were categorized as indeterminate were considered to have an infection for the purpose of analysis, for a prevalence of 22% in this cohort.
Conclusions: With respect to current diagnostic tests, rRNA-based RT-qPCR demonstrated 100% specificity and positive predictive value with a sensitivity equivalent to that of intraoperative culture. The RT-qPCR signal followed bacterial culture trends but exhibited detectable level for seven days after sterilization, allowing for the detection of infection after the antibiotic administration. These findings indicate that rRNA RT-qPCR is a sensitive and reliable test that retains the universal detection and speciation of DNA-based methods while functioning as a viability indicator.
Figures
References
-
- Clohisy JC, Calvert G, Tull F, McDonald D, Maloney WJ. Reasons for revision hip surgery: a retrospective review. Clin Orthop Relat Res. 2004;429:188-92 - PubMed
-
- Mulhall KJ, Ghomrawi HM, Scully S, Callaghan JJ, Saleh KJ. Current etiologies and modes of failure in total knee arthroplasty revision. Clin Orthop Relat Res. 2006;446:45-50 - PubMed
-
- Barrack RL, McClure JT, Burak CF, Clohisy JC, Parvizi J, Sharkey P. Revision total knee arthroplasty: the patient's perspective. Clin Orthop Relat Res. 2007;464:146-50 - PubMed
-
- Silva M, Tharani R, Schmalzried TP. Results of direct exchange or debridement of the infected total knee arthroplasty. Clin Orthop Relat Res. 2002;404:125-31 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

