Gene family size conservation is a good indicator of evolutionary rates
- PMID: 20194423
- PMCID: PMC2908708
- DOI: 10.1093/molbev/msq055
Gene family size conservation is a good indicator of evolutionary rates
Abstract
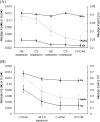
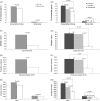
The evolution of duplicate genes has been a topic of broad interest. Here, we propose that the conservation of gene family size is a good indicator of the rate of sequence evolution and some other biological properties. By comparing the human-chimpanzee-macaque orthologous gene families with and without family size conservation, we demonstrate that genes with family size conservation evolve more slowly than those without family size conservation. Our results further demonstrate that both family expansion and contraction events may accelerate gene evolution, resulting in elevated evolutionary rates in the genes without family size conservation. In addition, we show that the duplicate genes with family size conservation evolve significantly more slowly than those without family size conservation. Interestingly, the median evolutionary rate of singletons falls in between those of the above two types of duplicate gene families. Our results thus suggest that the controversy on whether duplicate genes evolve more slowly than singletons can be resolved when family size conservation is taken into consideration. Furthermore, we also observe that duplicate genes with family size conservation have the highest level of gene expression/expression breadth, the highest proportion of essential genes, and the lowest gene compactness, followed by singletons and then by duplicate genes without family size conservation. Such a trend accords well with our observations of evolutionary rates. Our results thus point to the importance of family size conservation in the evolution of duplicate genes.
Figures
Similar articles
-
Origins and impact of constraints in evolution of gene families.Genome Res. 2006 Dec;16(12):1529-36. doi: 10.1101/gr.5346206. Epub 2006 Oct 19. Genome Res. 2006. PMID: 17053091 Free PMC article.
-
Rapid and asymmetric divergence of duplicate genes in the human gene coexpression network.BMC Bioinformatics. 2006 Jan 27;7:46. doi: 10.1186/1471-2105-7-46. BMC Bioinformatics. 2006. PMID: 16441884 Free PMC article.
-
Effect of duplicate genes on mouse genetic robustness: an update.Biomed Res Int. 2014;2014:758672. doi: 10.1155/2014/758672. Epub 2014 Jul 10. Biomed Res Int. 2014. PMID: 25110693 Free PMC article.
-
Concerted and birth-and-death evolution of multigene families.Annu Rev Genet. 2005;39:121-52. doi: 10.1146/annurev.genet.39.073003.112240. Annu Rev Genet. 2005. PMID: 16285855 Free PMC article. Review.
-
Molecular evolution of the chalcone synthase multigene family in the morning glory genome.Plant Mol Biol. 2000 Jan;42(1):79-92. Plant Mol Biol. 2000. PMID: 10688131 Review.
Cited by
-
The relationships among microRNA regulation, intrinsically disordered regions, and other indicators of protein evolutionary rate.Mol Biol Evol. 2011 Sep;28(9):2513-20. doi: 10.1093/molbev/msr068. Epub 2011 Mar 11. Mol Biol Evol. 2011. PMID: 21398349 Free PMC article.
-
Impacts of pretranscriptional DNA methylation, transcriptional transcription factor, and posttranscriptional microRNA regulations on protein evolutionary rate.Genome Biol Evol. 2014 Jun 12;6(6):1530-41. doi: 10.1093/gbe/evu124. Genome Biol Evol. 2014. PMID: 24923326 Free PMC article.
-
The impact of trans-regulation on the evolutionary rates of metazoan proteins.Nucleic Acids Res. 2013 Jul;41(13):6371-80. doi: 10.1093/nar/gkt349. Epub 2013 May 8. Nucleic Acids Res. 2013. PMID: 23658220 Free PMC article.
-
Whole Genome Duplications Shaped the Receptor Tyrosine Kinase Repertoire of Jawed Vertebrates.Genome Biol Evol. 2016 Jun 3;8(5):1600-13. doi: 10.1093/gbe/evw103. Genome Biol Evol. 2016. PMID: 27260203 Free PMC article.
-
Evolution and divergence of SBP-box genes in land plants.BMC Genomics. 2015 Oct 14;16:787. doi: 10.1186/s12864-015-1998-y. BMC Genomics. 2015. PMID: 26467431 Free PMC article.
References
-
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

