Personal genome sequencing: current approaches and challenges
- PMID: 20194435
- PMCID: PMC2827837
- DOI: 10.1101/gad.1864110
Personal genome sequencing: current approaches and challenges
Abstract
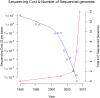
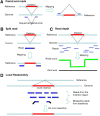
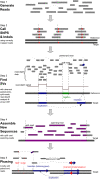
The revolution in DNA sequencing technologies has now made it feasible to determine the genome sequences of many individuals; i.e., "personal genomes." Genome sequences of cells and tissues from both normal and disease states have been determined. Using current approaches, whole human genome sequences are not typically assembled and determined de novo, but, instead, variations relative to a reference sequence are identified. We discuss the current state of personal genome sequencing, the main steps involved in determining a genome sequence (i.e., identifying single-nucleotide polymorphisms [SNPs] and structural variations [SVs], assembling new sequences, and phasing haplotypes), and the challenges and performance metrics for evaluating the accuracy of the reconstruction. Finally, we consider the possible individual and societal benefits of personal genome sequences.
Figures

References
-
- Archon X Prize Foundation. Archon X PRIZE competition guidelines. X Prize Foundation. 2006. http://genomics.xprize.org/files/downloads/genomics/Archon_X_PRIZE_for_G....
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
