miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia
- PMID: 20194440
- PMCID: PMC2827843
- DOI: 10.1101/gad.1856210
miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia
Abstract
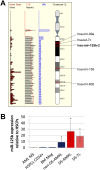
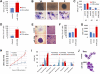
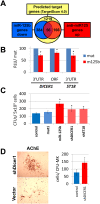
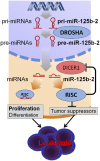
Children with trisomy 21/Down syndrome (DS) are at high risk to develop acute megakaryoblastic leukemia (DS-AMKL) and the related transient leukemia (DS-TL). The factors on human chromosome 21 (Hsa21) that confer this predisposing effect, especially in synergy with consistently mutated transcription factor GATA1 (GATA1s), remain poorly understood. Here, we investigated the role of Hsa21-encoded miR-125b-2, a microRNA (miRNA) overexpressed in DS-AMKL/TL, in hematopoiesis and leukemogenesis. We identified a function of miR-125b-2 in increasing proliferation and self-renewal of human and mouse megakaryocytic progenitors (MPs) and megakaryocytic/erythroid progenitors (MEPs). miR-125b-2 overexpression did not affect megakaryocytic and erythroid differentiation, but severely perturbed myeloid differentiation. The proproliferative effect of miR-125b-2 on MEPs accentuated the Gata1s mutation, whereas growth of DS-AMKL/TL cells was impaired upon miR-125b repression, suggesting synergism during leukemic transformation in GATA1s-mutated DS-AMKL/TL. Integrative transcriptome analysis of hematopoietic cells upon modulation of miR-125b expression levels uncovered a set of miR-125b target genes, including DICER1 and ST18 as direct targets. Gene Set Enrichment Analysis revealed that this target gene set is down-regulated in DS-AMKL patients highly expressing miR-125b. Thus, we propose miR-125b-2 as a positive regulator of megakaryopoiesis and an oncomiR involved in the pathogenesis of trisomy 21-associated megakaryoblastic leukemia.
Figures



Similar articles
-
The megakaryocytic transcription factor ARID3A suppresses leukemia pathogenesis.Blood. 2022 Feb 3;139(5):651-665. doi: 10.1182/blood.2021012231. Blood. 2022. PMID: 34570885 Free PMC article.
-
A unique role of GATA1s in Down syndrome acute megakaryocytic leukemia biology and therapy.PLoS One. 2011;6(11):e27486. doi: 10.1371/journal.pone.0027486. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22110660 Free PMC article.
-
Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis.Genes Dev. 2010 Aug 1;24(15):1659-72. doi: 10.1101/gad.1903410. Genes Dev. 2010. PMID: 20679399 Free PMC article.
-
Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome.Blood. 2009 Mar 19;113(12):2619-28. doi: 10.1182/blood-2008-11-163501. Epub 2009 Jan 12. Blood. 2009. PMID: 19139078 Free PMC article. Review.
-
Down syndrome and malignancies: a unique clinical relationship: a paper from the 2008 william beaumont hospital symposium on molecular pathology.J Mol Diagn. 2009 Sep;11(5):371-80. doi: 10.2353/jmoldx.2009.080132. J Mol Diagn. 2009. PMID: 19710397 Free PMC article. Review.
Cited by
-
MicroRNAs Associated with a Bad Prognosis in Acute Myeloid Leukemia and Their Impact on Macrophage Polarization.Biomedicines. 2024 Jan 7;12(1):121. doi: 10.3390/biomedicines12010121. Biomedicines. 2024. PMID: 38255226 Free PMC article. Review.
-
Clinical significance of microRNAs in chronic and acute human leukemia.Mol Cancer. 2016 May 14;15(1):37. doi: 10.1186/s12943-016-0518-2. Mol Cancer. 2016. PMID: 27179712 Free PMC article. Review.
-
Myt3 Mediates Laminin-V/Integrin-β1-Induced Islet-Cell Migration via Tgfbi.Mol Endocrinol. 2015 Sep;29(9):1254-68. doi: 10.1210/ME.2014-1387. Epub 2015 Jul 15. Mol Endocrinol. 2015. PMID: 26177052 Free PMC article.
-
CD33-targeting extracellular vesicles deliver antisense oligonucleotides against FLT3-ITD and miR-125b for specific treatment of acute myeloid leukaemia.Cell Prolif. 2022 Sep;55(9):e13255. doi: 10.1111/cpr.13255. Epub 2022 Jul 18. Cell Prolif. 2022. PMID: 35851970 Free PMC article.
-
Lin28: primal regulator of growth and metabolism in stem cells.Cell Stem Cell. 2013 Apr 4;12(4):395-406. doi: 10.1016/j.stem.2013.03.005. Cell Stem Cell. 2013. PMID: 23561442 Free PMC article. Review.
References
-
- Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. - PMC - PubMed
-
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
