Functional identification of two nonredundant Arabidopsis alpha(1,2)fucosyltransferases specific to arabinogalactan proteins
- PMID: 20194500
- PMCID: PMC2859526
- DOI: 10.1074/jbc.M110.102715
Functional identification of two nonredundant Arabidopsis alpha(1,2)fucosyltransferases specific to arabinogalactan proteins
Abstract
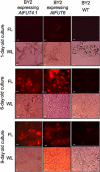
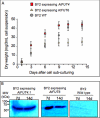
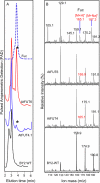
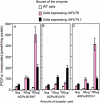
Virtually nothing is known about the mechanisms and enzymes responsible for the glycosylation of arabinogalactan proteins (AGPs). The glycosyltransferase 37 family contains plant-specific enzymes, which suggests involvement in plant-specific organs such as the cell wall. Our working hypothesis is that AtFUT4 and AtFUT6 genes encode alpha(1,2)fucosyltransferases (FUTs) for AGPs. Multiple lines of evidence support this hypothesis. First, overexpression of the two genes in tobacco BY2 cells, known to contain nonfucosylated AGPs, resulted in a staining of transgenic cells with eel lectin, which specifically binds to terminal alpha-linked fucose. Second, monosaccharide analysis by high pH anion exchange chromatography and electrospray ionization mass spectrometry indicated the presence of fucose in AGPs from transgenic cell lines but not in AGPs from wild type cells. Third, detergent extracts from microsomal membranes prepared from transgenic lines were able to fucosylate, in vitro, purified AGPs from BY2 wild type cells. Susceptibility of [(14)C]fucosylated AGPs to alpha(1,2)fucosidase, and not to alpha(1,3/4)fucosidase, indicated that an alpha(1,2) linkage is formed. Furthermore, dearabinosylated AGPs were not substrate acceptors for these enzymes, indicating that arabinosyl residues represent the fucosylation sites on these molecules. Testing of several polysaccharides, oligosaccharides, and glycoproteins as potential substrate acceptors in the fucosyl transfer reactions indicated that the two enzymes are specific for AGPs but are not functionally redundant because they differentially fucosylate certain AGPs. AtFUT4 and AtFUT6 are the first enzymes to be characterized for AGP glycosylation and further our understanding of cell wall biosynthesis.
Figures


Similar articles
-
Characterisation of FUT4 and FUT6 α-(1 → 2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity.PLoS One. 2014 Mar 25;9(3):e93291. doi: 10.1371/journal.pone.0093291. eCollection 2014. PLoS One. 2014. PMID: 24667545 Free PMC article.
-
Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis.J Exp Bot. 2013 Dec;64(18):5537-51. doi: 10.1093/jxb/ert321. Epub 2013 Oct 14. J Exp Bot. 2013. PMID: 24127514 Free PMC article.
-
AtFUT4 and AtFUT6 Are Arabinofuranose-Specific Fucosyltransferases.Front Plant Sci. 2021 Feb 9;12:589518. doi: 10.3389/fpls.2021.589518. eCollection 2021. Front Plant Sci. 2021. PMID: 33633757 Free PMC article.
-
Fucosylation in Urological Cancers.Int J Mol Sci. 2021 Dec 11;22(24):13333. doi: 10.3390/ijms222413333. Int J Mol Sci. 2021. PMID: 34948129 Free PMC article. Review.
-
Back to the future with the AGP-Ca2+ flux capacitor.Ann Bot. 2014 Oct;114(6):1069-85. doi: 10.1093/aob/mcu161. Epub 2014 Aug 19. Ann Bot. 2014. PMID: 25139429 Free PMC article. Review.
Cited by
-
Cyclin-Dependent Kinase Inhibitor Gene TaICK1 acts as a Potential Contributor to Wheat Male Sterility induced by a Chemical Hybridizing Agent.Int J Mol Sci. 2020 Apr 2;21(7):2468. doi: 10.3390/ijms21072468. Int J Mol Sci. 2020. PMID: 32252420 Free PMC article.
-
Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae.Ann Bot. 2014 Oct;114(6):1217-36. doi: 10.1093/aob/mcu171. Epub 2014 Sep 9. Ann Bot. 2014. PMID: 25204387 Free PMC article.
-
A Pipeline towards the Biochemical Characterization of the Arabidopsis GT14 Family.Int J Mol Sci. 2021 Jan 29;22(3):1360. doi: 10.3390/ijms22031360. Int J Mol Sci. 2021. PMID: 33572987 Free PMC article.
-
Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides.Plant Physiol. 2012 Oct;160(2):653-66. doi: 10.1104/pp.112.202309. Epub 2012 Aug 13. Plant Physiol. 2012. PMID: 22891237 Free PMC article.
-
Characterisation of FUT4 and FUT6 α-(1 → 2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity.PLoS One. 2014 Mar 25;9(3):e93291. doi: 10.1371/journal.pone.0093291. eCollection 2014. PLoS One. 2014. PMID: 24667545 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

