Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34
- PMID: 20194622
- PMCID: PMC2863694
- DOI: 10.1128/MCB.01094-09
Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34
Abstract
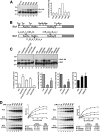
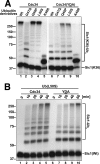
Ubiquitin (Ub)-conjugating enzymes (E2s) and ubiquitin ligases (E3s) catalyze the attachment of Ub to lysine residues in substrates and Ub during monoubiquitination and polyubiquitination. Lysine selection is important for the generation of diverse substrate-Ub structures, which provides versatility to this pathway in the targeting of proteins to different fates. The mechanisms of lysine selection remain poorly understood, with previous studies suggesting that the ubiquitination site(s) is selected by the E2/E3-mediated positioning of a lysine(s) toward the E2/E3 active site. By studying the polyubiquitination of Sic1 by the E2 protein Cdc34 and the RING E3 Skp1/Cul1/F-box (SCF) protein, we now demonstrate that in addition to E2/E3-mediated positioning, proximal amino acids surrounding the lysine residues in Sic1 and Ub are critical for ubiquitination. This mechanism is linked to key residues composing the catalytic core of Cdc34 and independent of SCF. Changes to these core residues altered the lysine preference of Cdc34 and specified whether this enzyme monoubiquitinated or polyubiquitinated Sic1. These new findings indicate that compatibility between amino acids surrounding acceptor lysine residues and key amino acids in the catalytic core of ubiquitin-conjugating enzymes is an important mechanism for lysine selection during ubiquitination.
Figures





Similar articles
-
Molecular and structural insight into lysine selection on substrate and ubiquitin lysine 48 by the ubiquitin-conjugating enzyme Cdc34.Cell Cycle. 2013 Jun 1;12(11):1732-44. doi: 10.4161/cc.24818. Epub 2013 May 8. Cell Cycle. 2013. PMID: 23656784 Free PMC article.
-
Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34.Cell. 2005 Dec 16;123(6):1107-20. doi: 10.1016/j.cell.2005.09.033. Cell. 2005. PMID: 16360039
-
Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression.Biochem J. 2007 Aug 1;405(3):569-81. doi: 10.1042/BJ20061812. Biochem J. 2007. PMID: 17461777 Free PMC article.
-
Structural basis of generic versus specific E2-RING E3 interactions in protein ubiquitination.Protein Sci. 2019 Oct;28(10):1758-1770. doi: 10.1002/pro.3690. Epub 2019 Aug 23. Protein Sci. 2019. PMID: 31340062 Free PMC article. Review.
-
Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination.FEBS J. 2009 Oct;276(19):5381-9. doi: 10.1111/j.1742-4658.2009.07249.x. Epub 2009 Aug 27. FEBS J. 2009. PMID: 19712108 Free PMC article. Review.
Cited by
-
Tripartite degrons confer diversity and specificity on regulated protein degradation in the ubiquitin-proteasome system.Nat Commun. 2016 Jan 6;7:10239. doi: 10.1038/ncomms10239. Nat Commun. 2016. PMID: 26732515 Free PMC article.
-
The loop-less tmCdc34 E2 mutant defective in polyubiquitination in vitro and in vivo supports yeast growth in a manner dependent on Ubp14 and Cka2.Cell Div. 2011 Mar 31;6:7. doi: 10.1186/1747-1028-6-7. Cell Div. 2011. PMID: 21453497 Free PMC article.
-
Increased CSN5 expression enhances the sensitivity to lenalidomide in multiple myeloma cells.iScience. 2024 Nov 15;27(12):111399. doi: 10.1016/j.isci.2024.111399. eCollection 2024 Dec 20. iScience. 2024. PMID: 39687025 Free PMC article.
-
SCFCdc4 enables mating type switching in yeast by cyclin-dependent kinase-mediated elimination of the Ash1 transcriptional repressor.Mol Cell Biol. 2011 Feb;31(3):584-98. doi: 10.1128/MCB.00845-10. Epub 2010 Nov 22. Mol Cell Biol. 2011. PMID: 21098119 Free PMC article.
-
Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3.Elife. 2013 Aug 8;2:e00828. doi: 10.7554/eLife.00828. Elife. 2013. PMID: 23936628 Free PMC article.
References
-
- Banerjee, A., L. Gregori, Y. Xu, and V. Chau. 1993. The bacterially expressed yeast cdc34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J. Biol. Chem. 268:5668-5675. - PubMed
-
- Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. - PubMed
-
- Block, K., T. Boyer, and P. Yew. 2001. Phosphorylation of the human ubiquitin-conjugating enzyme, Cdc34, by casein kinase 2. J. Biol. Chem. 276:41049-41058. - PubMed
-
- Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. - PubMed
-
- Deffenbaugh, A., K. Scaglione, L. Zhang, J. Moore, T. Buranda, L. Sklar, and D. Skowyra. 2003. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCFcdc4-bound substrate SIC1. Cell 114:611-622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
