Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells
- PMID: 20194747
- PMCID: PMC2841923
- DOI: 10.1073/pnas.1000896107
Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells
Abstract
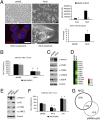
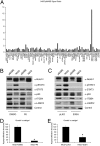
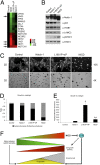
Aberrant activation of Notch receptors has been implicated in breast cancer; however, the mechanisms contributing to Notch-dependent transformation remain elusive because Notch displays dichotomous functional activities, promoting both proliferation and growth arrest. We investigated the cellular basis for the heterogeneous responses to Notch pathway activation in 3D cultures of MCF-10A mammary epithelial cells. Expression of a constitutively active Notch-1 intracellular domain (NICD) was found to induce two distinct types of 3D structures: large, hyperproliferative structures and small, growth-arrested structures with reduced cell-to-matrix adhesion. Interestingly, we found that these heterogeneous phenotypes reflect differences in Notch pathway activation levels; high Notch activity caused down-regulation of multiple matrix-adhesion genes and inhibition of proliferation, whereas low Notch activity maintained matrix adhesion and provoked a strong hyperproliferative response. Moreover, microarray analyses implicated NICD-induced p63 down-regulation in loss of matrix adhesion. In addition, a reverse-phase protein array-based analysis and subsequent loss-of-function studies identified STAT3 as a dominant downstream mediator of the NICD-induced outgrowth. These results indicate that the phenotypic responses to Notch are determined by the dose of pathway activation; and this dose affects the balance between growth-stimulative and growth-suppressive effects. This unique feature of Notch signaling provides insights into mechanisms that contribute to the dichotomous effects of Notch during development and tumorigenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. - PubMed
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Radtke F, Raj K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. - PubMed
-
- Rizzo P, et al. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. - PubMed
-
- Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

