Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin
- PMID: 20194754
- PMCID: PMC2841909
- DOI: 10.1073/pnas.0913485107
Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin
Abstract
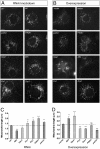
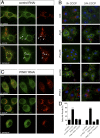
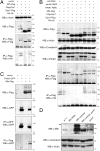
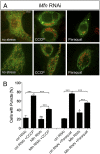
Loss of the E3 ubiquitin ligase Parkin causes early onset Parkinson's disease, a neurodegenerative disorder of unknown etiology. Parkin has been linked to multiple cellular processes including protein degradation, mitochondrial homeostasis, and autophagy; however, its precise role in pathogenesis is unclear. Recent evidence suggests that Parkin is recruited to damaged mitochondria, possibly affecting mitochondrial fission and/or fusion, to mediate their autophagic turnover. The precise mechanism of recruitment and the ubiquitination target are unclear. Here we show in Drosophila cells that PINK1 is required to recruit Parkin to dysfunctional mitochondria and promote their degradation. Furthermore, PINK1 and Parkin mediate the ubiquitination of the profusion factor Mfn on the outer surface of mitochondria. Loss of Drosophila PINK1 or parkin causes an increase in Mfn abundance in vivo and concomitant elongation of mitochondria. These findings provide a molecular mechanism by which the PINK1/Parkin pathway affects mitochondrial fission/fusion as suggested by previous genetic interaction studies. We hypothesize that Mfn ubiquitination may provide a mechanism by which terminally damaged mitochondria are labeled and sequestered for degradation by autophagy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gasser T. Molecular pathogenesis of Parkinson disease: Insights from genetic studies. Expert Rev Mol Med. 2009;11:e22. - PubMed
-
- Silvestri L, et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–3492. - PubMed
-
- Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. - PubMed
-
- Büeler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson's disease. Exp Neurol. 2009;218:235–246. - PubMed
-
- Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

