Evolution of species-specific promoter-associated mechanisms for protecting chromosome ends by Drosophila Het-A telomeric transposons
- PMID: 20194755
- PMCID: PMC2841908
- DOI: 10.1073/pnas.1000612107
Evolution of species-specific promoter-associated mechanisms for protecting chromosome ends by Drosophila Het-A telomeric transposons
Abstract
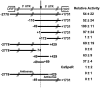
The non-LTR retrotransposons forming Drosophila telomeres constitute a robust mechanism for telomere maintenance, one which has persisted since before separation of the extant Drosophila species. These elements in D. melanogaster differ from nontelomeric retrotransposons in ways that give insight into general telomere biology. Here, we analyze telomere-specific retrotransposons from D. virilis, separated from D. melanogaster by 40 to 60 million years, to evaluate the evolutionary divergence of their telomeric traits. The telomeric retrotransposon HeT-A from D. melanogaster has an unusual promoter near its 3' terminus that drives not the element in which it resides, but the adjacent downstream element in a head-to-tail array. An obvious benefit of this promoter is that it adds nonessential sequence to the 5' end of each transcript, which is reverse transcribed and added to the chromosome. Because the 5' end of each newly transposed element forms the end of the chromosome until another element transposes onto it, this nonessential sequence can buffer erosion of sequence essential for HeT-A. Surprisingly, we have now found that HeT-A in D. virilis has a promoter typical of non-LTR retrotransposons. This promoter adds no buffering sequence; nevertheless, the complete 5' end of the element persists in telomere arrays, necessitating a more precise processing of the extreme end of the telomere in D. virilis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

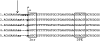
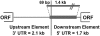
Similar articles
-
Evolution of diverse mechanisms for protecting chromosome ends by Drosophila TART telomere retrotransposons.Proc Natl Acad Sci U S A. 2010 Dec 7;107(49):21052-7. doi: 10.1073/pnas.1015926107. Epub 2010 Nov 18. Proc Natl Acad Sci U S A. 2010. PMID: 21088221 Free PMC article.
-
Unusual features of the Drosophila melanogaster telomere transposable element HeT-A are conserved in Drosophila yakuba telomere elements.Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3770-5. doi: 10.1073/pnas.95.7.3770. Proc Natl Acad Sci U S A. 1998. PMID: 9520442 Free PMC article.
-
Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs.Cell. 1997 Mar 7;88(5):647-55. doi: 10.1016/s0092-8674(00)81907-8. Cell. 1997. PMID: 9054504
-
Drosophila telomere transposons: genetically active elements in heterochromatin.Genetica. 2000;109(1-2):45-52. doi: 10.1023/a:1026540301503. Genetica. 2000. PMID: 11293794 Review.
-
Drosophila telomeres: A variation on the telomerase theme.Fly (Austin). 2008 May-Jun;2(3):101-10. doi: 10.4161/fly.6393. Epub 2008 May 4. Fly (Austin). 2008. PMID: 18820466 Review.
Cited by
-
Retrotransposons that maintain chromosome ends.Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20317-24. doi: 10.1073/pnas.1100278108. Epub 2011 Aug 5. Proc Natl Acad Sci U S A. 2011. PMID: 21821789 Free PMC article.
-
Evolution of diverse mechanisms for protecting chromosome ends by Drosophila TART telomere retrotransposons.Proc Natl Acad Sci U S A. 2010 Dec 7;107(49):21052-7. doi: 10.1073/pnas.1015926107. Epub 2010 Nov 18. Proc Natl Acad Sci U S A. 2010. PMID: 21088221 Free PMC article.
-
Adapting to life at the end of the line: How Drosophila telomeric retrotransposons cope with their job.Mob Genet Elements. 2011 Jul;1(2):128-134. doi: 10.4161/mge.1.2.16914. Epub 2011 Jul 1. Mob Genet Elements. 2011. PMID: 22016861 Free PMC article.
-
Differential maintenance of DNA sequences in telomeric and centromeric heterochromatin.Genetics. 2011 Jan;187(1):51-60. doi: 10.1534/genetics.110.122994. Epub 2010 Nov 1. Genetics. 2011. PMID: 21041555 Free PMC article.
-
Drosophila: Retrotransposons Making up Telomeres.Viruses. 2017 Jul 19;9(7):192. doi: 10.3390/v9070192. Viruses. 2017. PMID: 28753967 Free PMC article. Review.
References
-
- Pardue ML, DeBaryshe PG. In: Origin and Evolution of Telomeres. Nosek J, Tomaska L, editors. Austin, TX: Landes Bioscience; 2008. pp. 27–44.
-
- Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

