Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124
- PMID: 20194791
- PMCID: PMC2841876
- DOI: 10.1073/pnas.0909141107
Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124
Abstract
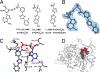
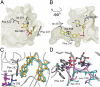
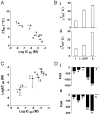
Firefly luciferase (FLuc), an ATP-dependent bioluminescent reporter enzyme, is broadly used in chemical biology and drug discovery assays. PTC124 (Ataluren; (3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]benzoic acid) discovered in an FLuc-based assay targeting nonsense codon suppression, is an unusually potent FLuc-inhibitor. Paradoxically, PTC124 and related analogs increase cellular FLuc activity levels by posttranslational stabilization. In this study, we show that FLuc inhibition and stabilization is the result of an inhibitory product formed during the FLuc-catalyzed reaction between its natural substrate, ATP, and PTC124. A 2.0 A cocrystal structure revealed the inhibitor to be the acyl-AMP mixed-anhydride adduct PTC124-AMP, which was subsequently synthesized and shown to be a high-affinity multisubstrate adduct inhibitor (MAI; K(D) = 120 pM) of FLuc. Biochemical assays, liquid chromatography/mass spectrometry, and near-attack conformer modeling demonstrate that formation of this novel MAI is absolutely dependent upon the precise positioning and reactivity of a key meta-carboxylate of PTC124 within the FLuc active site. We also demonstrate that the inhibitory activity of PTC124-AMP is relieved by free coenzyme A, a component present at high concentrations in luciferase detection reagents used for cell-based assays. This explains why PTC124 can appear to increase, instead of inhibit, FLuc activity in cell-based reporter gene assays. To our knowledge, this is an unusual example in which the "off-target" effect of a small molecule is mediated by an MAI mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.Chem Biol. 2012 Aug 24;19(8):1060-72. doi: 10.1016/j.chembiol.2012.07.015. Chem Biol. 2012. PMID: 22921073 Free PMC article.
-
Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression.Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3585-90. doi: 10.1073/pnas.0813345106. Epub 2009 Feb 10. Proc Natl Acad Sci U S A. 2009. PMID: 19208811 Free PMC article.
-
Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology.Chem Biol. 2010 Jun 25;17(6):646-57. doi: 10.1016/j.chembiol.2010.05.012. Chem Biol. 2010. PMID: 20609414 Free PMC article. Review.
-
A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays.PLoS Biol. 2013;11(6):e1001593. doi: 10.1371/journal.pbio.1001593. Epub 2013 Jun 25. PLoS Biol. 2013. PMID: 23824517 Free PMC article.
-
Firefly luciferase inhibition.J Photochem Photobiol B. 2010 Oct 5;101(1):1-8. doi: 10.1016/j.jphotobiol.2010.06.015. Epub 2010 Jul 3. J Photochem Photobiol B. 2010. PMID: 20655239 Review.
Cited by
-
PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS.J Med Chem. 2015 Mar 12;58(5):2091-113. doi: 10.1021/jm5019093. Epub 2015 Feb 21. J Med Chem. 2015. PMID: 25634295 Free PMC article.
-
Improved Synthesis of Biotinol-5'-AMP: Implications for Antibacterial Discovery.ACS Med Chem Lett. 2014 Dec 11;6(2):216-20. doi: 10.1021/ml500475n. eCollection 2015 Feb 12. ACS Med Chem Lett. 2014. PMID: 25699152 Free PMC article.
-
Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.Chem Biol. 2012 Aug 24;19(8):1060-72. doi: 10.1016/j.chembiol.2012.07.015. Chem Biol. 2012. PMID: 22921073 Free PMC article.
-
Targeting Nonsense: Optimization of 1,2,4-Oxadiazole TRIDs to Rescue CFTR Expression and Functionality in Cystic Fibrosis Cell Model Systems.Int J Mol Sci. 2020 Sep 3;21(17):6420. doi: 10.3390/ijms21176420. Int J Mol Sci. 2020. PMID: 32899265 Free PMC article.
-
Clinical potential of ataluren in the treatment of Duchenne muscular dystrophy.Degener Neurol Neuromuscul Dis. 2016 May 13;6:37-48. doi: 10.2147/DNND.S71808. eCollection 2016. Degener Neurol Neuromuscul Dis. 2016. PMID: 30050367 Free PMC article. Review.
References
-
- Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5(1):127–136. - PubMed
-
- Roda A, Guardigli M, Michelini E, Mirasoli M. Bioluminescence in analytical chemistry and in vivo imaging. TRAC-Trend Anal Chem. 2009;28(3):307–322.
-
- Auld DS, et al. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008;51(8):2372–2386. - PubMed
-
- Heitman LH, et al. False positives in a reporter gene assay: Identification and synthesis of substituted N-pyridin-2-ylbenzamides as competitive inhibitors of firefly luciferase. J Med Chem. 2008;51(15):4724–4729. - PubMed
-
- Thompson JF, et al. Mutation of a protease-sensitive region in firefly luciferase alters light emission properties. J Biol Chem. 1997;272(30):18766–18771. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

