Condensins promote coorientation of sister chromatids during meiosis I in budding yeast
- PMID: 20194961
- PMCID: PMC2870976
- DOI: 10.1534/genetics.110.115139
Condensins promote coorientation of sister chromatids during meiosis I in budding yeast
Abstract
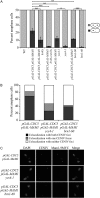
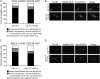
The condensin complex is a key determinant of higher-ordered chromosome structure. We show here that the complex is also important for the correct alignment of chromosomes on the meiosis I spindle. Unlike during mitosis and meiosis II, when sister chromatids attach to microtubules emanating from opposite spindle poles (biorientation), accurate meiosis I chromosome segregation requires that sister chromatids attach to microtubules emanating from the same spindle pole (co-orientation). The monopolin complex, consisting of Lrs4, Csm1, and the meiosis-specific component Mam1, brings about meiosis I co-orientation. We find that in the absence of functional condensin complexes, a fraction of sister kinetochores biorient on the meiosis I spindle and association of the monopolin complex subunit Mam1 with kinetochores is decreased. Our studies uncover a new locus-specific effect of the condensin complex.
Figures





Similar articles
-
Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex.Cell. 2007 Feb 9;128(3):477-90. doi: 10.1016/j.cell.2006.12.040. Cell. 2007. PMID: 17289568 Free PMC article.
-
Monopolin subunit Csm1 associates with MIND complex to establish monopolar attachment of sister kinetochores at meiosis I.PLoS Genet. 2013;9(7):e1003610. doi: 10.1371/journal.pgen.1003610. Epub 2013 Jul 4. PLoS Genet. 2013. PMID: 23861669 Free PMC article.
-
Molecular architecture of the yeast monopolin complex.Cell Rep. 2012 Jun 28;1(6):583-9. doi: 10.1016/j.celrep.2012.05.012. Epub 2012 Jun 21. Cell Rep. 2012. PMID: 22813733 Free PMC article.
-
Deciphering condensin action during chromosome segregation.Trends Cell Biol. 2011 Sep;21(9):552-9. doi: 10.1016/j.tcb.2011.06.003. Epub 2011 Jul 15. Trends Cell Biol. 2011. PMID: 21763138 Review.
-
Condensin in Chromatid Cohesion and Segregation.Cytogenet Genome Res. 2015;147(4):212-6. doi: 10.1159/000444868. Epub 2016 Mar 22. Cytogenet Genome Res. 2015. PMID: 26998746 Review.
Cited by
-
Roles of cohesin and condensin in chromosome dynamics during mammalian meiosis.J Reprod Dev. 2013 Oct;59(5):431-6. doi: 10.1262/jrd.2013-068. J Reprod Dev. 2013. PMID: 24162807 Free PMC article. Review.
-
Condensin: crafting the chromosome landscape.Chromosoma. 2013 Jun;122(3):175-90. doi: 10.1007/s00412-013-0405-1. Epub 2013 Apr 2. Chromosoma. 2013. PMID: 23546018 Review.
-
The negatively charged carboxy-terminal tail of β-tubulin promotes proper chromosome segregation.Mol Biol Cell. 2016 Jun 1;27(11):1786-96. doi: 10.1091/mbc.E15-05-0300. Epub 2016 Apr 6. Mol Biol Cell. 2016. PMID: 27053662 Free PMC article.
-
The budding-yeast RWD protein Csm1 scaffolds diverse protein complexes through a conserved structural mechanism.Protein Sci. 2018 Dec;27(12):2094-2100. doi: 10.1002/pro.3515. Epub 2018 Nov 5. Protein Sci. 2018. PMID: 30252178 Free PMC article.
-
Evidence of Zip1 Promoting Sister Kinetochore Mono-orientation During Meiosis in Budding Yeast.G3 (Bethesda). 2018 Nov 6;8(11):3691-3701. doi: 10.1534/g3.118.200469. G3 (Bethesda). 2018. PMID: 30254179 Free PMC article.
References
-
- Amon, A., 2002. Synchronization procedures. Methods Enzymol. 351 457–467. - PubMed
-
- Chelysheva, L., S. Diallo, D. Vezon, G. Gendrot, N. Vrielynck et al., 2005. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118 4621–4632. - PubMed
-
- Clyne, R. K., V. L. Katis, L. Jessop, K. R. Benjamin, I. Herskowitz et al., 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell. Biol. 5 480–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

