The nucleosome remodeling factor ISWI functionally interacts with an evolutionarily conserved network of cellular factors
- PMID: 20194965
- PMCID: PMC2870949
- DOI: 10.1534/genetics.110.114256
The nucleosome remodeling factor ISWI functionally interacts with an evolutionarily conserved network of cellular factors
Abstract
ISWI is an evolutionarily conserved ATP-dependent chromatin remodeling factor playing central roles in DNA replication, RNA transcription, and chromosome organization. The variety of biological functions dependent on ISWI suggests that its activity could be highly regulated. Our group has previously isolated and characterized new cellular activities that positively regulate ISWI in Drosophila melanogaster. To identify factors that antagonize ISWI activity we developed a novel in vivo eye-based assay to screen for genetic suppressors of ISWI. Our screen revealed that ISWI interacts with an evolutionarily conserved network of cellular and nuclear factors that escaped previous genetic and biochemical analyses.
Figures



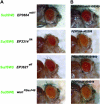
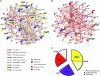

References
-
- Andersen, E. C., X. Lu and H. R. Horvitz, 2006. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133 2695–2704. - PubMed
-
- Armstrong, J. A., A. S. Sperling, R. Deuring, L. Manning, S. L. Moseley et al., 2005. Genetic screens for enhancers of brahma reveal functional interactions between the BRM chromatin-remodeling complex and the delta-notch signal transduction pathway in Drosophila. Genetics 170 1761–1774. - PMC - PubMed
-
- Badenhorst, P., S. Harrison and A. Travers, 1996. End of the line? Tramtrack and cell fate determination in Drosophila. Genes Cells 1 707–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

