A primer on metagenomics
- PMID: 20195499
- PMCID: PMC2829047
- DOI: 10.1371/journal.pcbi.1000667
A primer on metagenomics
Abstract
Metagenomics is a discipline that enables the genomic study of uncultured microorganisms. Faster, cheaper sequencing technologies and the ability to sequence uncultured microbes sampled directly from their habitats are expanding and transforming our view of the microbial world. Distilling meaningful information from the millions of new genomic sequences presents a serious challenge to bioinformaticians. In cultured microbes, the genomic data come from a single clone, making sequence assembly and annotation tractable. In metagenomics, the data come from heterogeneous microbial communities, sometimes containing more than 10,000 species, with the sequence data being noisy and partial. From sampling, to assembly, to gene calling and function prediction, bioinformatics faces new demands in interpreting voluminous, noisy, and often partial sequence data. Although metagenomics is a relative newcomer to science, the past few years have seen an explosion in computational methods applied to metagenomic-based research. It is therefore not within the scope of this article to provide an exhaustive review. Rather, we provide here a concise yet comprehensive introduction to the current computational requirements presented by metagenomics, and review the recent progress made. We also note whether there is software that implements any of the methods presented here, and briefly review its utility. Nevertheless, it would be useful if readers of this article would avail themselves of the comment section provided by this journal, and relate their own experiences. Finally, the last section of this article provides a few representative studies illustrating different facets of recent scientific discoveries made using metagenomics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

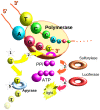
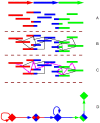
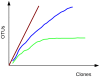
References
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
-
- Berg R. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. - PubMed
-
- Collins FS, McKusick VA. Implications of the human genome project for medical science. JAMA. 2001;285:540–544. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

