Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals
- PMID: 20195503
- PMCID: PMC2829051
- DOI: 10.1371/journal.ppat.1000781
Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals
Abstract
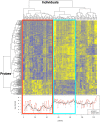
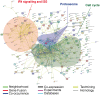
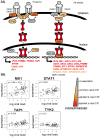
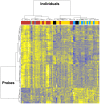
There is great interindividual variability in HIV-1 viral setpoint after seroconversion, some of which is known to be due to genetic differences among infected individuals. Here, our focus is on determining, genome-wide, the contribution of variable gene expression to viral control, and to relate it to genomic DNA polymorphism. RNA was extracted from purified CD4+ T-cells from 137 HIV-1 seroconverters, 16 elite controllers, and 3 healthy blood donors. Expression levels of more than 48,000 mRNA transcripts were assessed by the Human-6 v3 Expression BeadChips (Illumina). Genome-wide SNP data was generated from genomic DNA using the HumanHap550 Genotyping BeadChip (Illumina). We observed two distinct profiles with 260 genes differentially expressed depending on HIV-1 viral load. There was significant upregulation of expression of interferon stimulated genes with increasing viral load, including genes of the intrinsic antiretroviral defense. Upon successful antiretroviral treatment, the transcriptome profile of previously viremic individuals reverted to a pattern comparable to that of elite controllers and of uninfected individuals. Genome-wide evaluation of cis-acting SNPs identified genetic variants modulating expression of 190 genes. Those were compared to the genes whose expression was found associated with viral load: expression of one interferon stimulated gene, OAS1, was found to be regulated by a SNP (rs3177979, p = 4.9E-12); however, we could not detect an independent association of the SNP with viral setpoint. Thus, this study represents an attempt to integrate genome-wide SNP signals with genome-wide expression profiles in the search for biological correlates of HIV-1 control. It underscores the paradox of the association between increasing levels of viral load and greater expression of antiviral defense pathways. It also shows that elite controllers do not have a fully distinctive mRNA expression pattern in CD4+ T cells. Overall, changes in global RNA expression reflect responses to viral replication rather than a mechanism that might explain viral control.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS ONE. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. - DOI - PMC - PubMed
-
- Limou S, Le CS, Coulonges C, Carpentier W, Dina C, et al. Genomewide Association Study of an AIDS-Nonprogression Cohort Emphasizes the Role Played by HLA Genes (ANRS Genomewide Association Study 02). J Infect Dis. 2009;199:419–426. - PubMed
-
- Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, et al. Common Genetic Variation and the Control of HIV-1 in Humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

