A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology
- PMID: 20195509
- PMCID: PMC2829057
- DOI: 10.1371/journal.ppat.1000784
A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology
Abstract
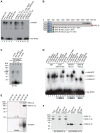
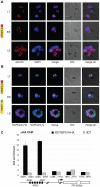
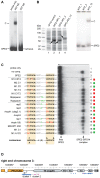
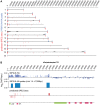
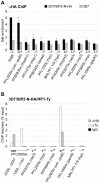
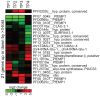
The heterochromatic environment and physical clustering of chromosome ends at the nuclear periphery provide a functional and structural framework for antigenic variation and evolution of subtelomeric virulence gene families in the malaria parasite Plasmodium falciparum. While recent studies assigned important roles for reversible histone modifications, silent information regulator 2 and heterochromatin protein 1 (PfHP1) in epigenetic control of variegated expression, factors involved in the recruitment and organization of subtelomeric heterochromatin remain unknown. Here, we describe the purification and characterization of PfSIP2, a member of the ApiAP2 family of putative transcription factors, as the unknown nuclear factor interacting specifically with cis-acting SPE2 motif arrays in subtelomeric domains. Interestingly, SPE2 is not bound by the full-length protein but rather by a 60kDa N-terminal domain, PfSIP2-N, which is released during schizogony. Our experimental re-definition of the SPE2/PfSIP2-N interaction highlights the strict requirement of both adjacent AP2 domains and a conserved bipartite SPE2 consensus motif for high-affinity binding. Genome-wide in silico mapping identified 777 putative binding sites, 94% of which cluster in heterochromatic domains upstream of subtelomeric var genes and in telomere-associated repeat elements. Immunofluorescence and chromatin immunoprecipitation (ChIP) assays revealed co-localization of PfSIP2-N with PfHP1 at chromosome ends. Genome-wide ChIP demonstrated the exclusive binding of PfSIP2-N to subtelomeric SPE2 landmarks in vivo but not to single chromosome-internal sites. Consistent with this specialized distribution pattern, PfSIP2-N over-expression has no effect on global gene transcription. Hence, contrary to the previously proposed role for this factor in gene activation, our results provide strong evidence for the first time for the involvement of an ApiAP2 factor in heterochromatin formation and genome integrity. These findings are highly relevant for our understanding of chromosome end biology and variegated expression in P. falciparum and other eukaryotes, and for the future analysis of the role of ApiAP2-DNA interactions in parasite biology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Evidence that Plasmodium falciparum chromosome end clusters are cross-linked by protein and are the sites of both virulence gene silencing and activation.Mol Microbiol. 2006 Oct;62(1):72-83. doi: 10.1111/j.1365-2958.2006.05364.x. Epub 2006 Aug 30. Mol Microbiol. 2006. PMID: 16942599
-
A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation.EMBO J. 2002 Feb 15;21(4):815-24. doi: 10.1093/emboj/21.4.815. EMBO J. 2002. PMID: 11847128 Free PMC article.
-
Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors.PLoS Pathog. 2009 Sep;5(9):e1000569. doi: 10.1371/journal.ppat.1000569. Epub 2009 Sep 4. PLoS Pathog. 2009. PMID: 19730695 Free PMC article.
-
Impact of chromosome ends on the biology and virulence of Plasmodium falciparum.Mol Biochem Parasitol. 2013 Feb;187(2):121-8. doi: 10.1016/j.molbiopara.2013.01.003. Epub 2013 Jan 24. Mol Biochem Parasitol. 2013. PMID: 23354131 Review.
-
Mutually exclusive var gene expression in the malaria parasite: multiple layers of regulation.Trends Parasitol. 2008 Oct;24(10):455-61. doi: 10.1016/j.pt.2008.07.005. Epub 2008 Sep 2. Trends Parasitol. 2008. PMID: 18771955 Review.
Cited by
-
Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites.Elife. 2015 Jul 15;4:e06974. doi: 10.7554/eLife.06974. Elife. 2015. PMID: 26175406 Free PMC article.
-
Plasmodium falciparum Development from Gametocyte to Oocyst: Insight from Functional Studies.Microorganisms. 2023 Jul 31;11(8):1966. doi: 10.3390/microorganisms11081966. Microorganisms. 2023. PMID: 37630530 Free PMC article. Review.
-
Antibody Biomarkers Associated with Sterile Protection Induced by Controlled Human Malaria Infection under Chloroquine Prophylaxis.mSphere. 2019 Feb 20;4(1):e00027-19. doi: 10.1128/mSphereDirect.00027-19. mSphere. 2019. PMID: 30787114 Free PMC article.
-
PfSWIB, a potential chromatin regulator for var gene regulation and parasite development in Plasmodium falciparum.Parasit Vectors. 2020 Feb 4;13(1):48. doi: 10.1186/s13071-020-3918-5. Parasit Vectors. 2020. PMID: 32019597 Free PMC article.
-
Regulation of Sexual Commitment and Gametocytogenesis in Malaria Parasites.Annu Rev Microbiol. 2018 Sep 8;72:501-519. doi: 10.1146/annurev-micro-090817-062712. Epub 2018 Jul 5. Annu Rev Microbiol. 2018. PMID: 29975590 Free PMC article. Review.
References
-
- Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin Genet Dev. 1997;7:822–828. - PubMed
-
- Taddei A, Hediger F, Neumann FR, Gasser SM. The function of nuclear architecture: a genetic approach. Annu Rev Genet. 2004;38:305–345. - PubMed
-
- Louis EJ, Vershinin AV. Chromosome ends: different sequences may provide conserved functions. Bioessays. 2005;27:685–697. - PubMed
-
- Tham WH, Zakian VA. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002;21:512–521. - PubMed
-
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

