Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells
- PMID: 20195511
- PMCID: PMC2829059
- DOI: 10.1371/journal.pgen.1000855
Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells
Abstract
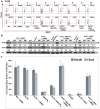
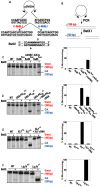
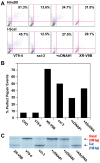
The repair of DNA double-strand breaks (DSBs) is critical for the maintenance of genomic integrity and viability for all organisms. Mammals have evolved at least two genetically discrete ways to mediate DNA DSB repair: homologous recombination (HR) and non-homologous end joining (NHEJ). In mammalian cells, most DSBs are preferentially repaired by NHEJ. Recent work has demonstrated that NHEJ consists of at least two sub-pathways-the main Ku heterodimer-dependent or "classic" NHEJ (C-NHEJ) pathway and an "alternative" NHEJ (A-NHEJ) pathway, which usually generates microhomology-mediated signatures at repair junctions. In our study, recombinant adeno-associated virus knockout vectors were utilized to construct a series of isogenic human somatic cell lines deficient in the core C-NHEJ factors (Ku, DNA-PK(cs), XLF, and LIGIV), and the resulting cell lines were characterized for their ability to carry out DNA DSB repair. The absence of DNA-PK(cs), XLF, or LIGIV resulted in cell lines that were profoundly impaired in DNA DSB repair activity. Unexpectedly, Ku86-null cells showed wild-type levels of DNA DSB repair activity that was dominated by microhomology joining events indicative of A-NHEJ. Importantly, A-NHEJ DNA DSB repair activity could also be efficiently de-repressed in LIGIV-null and DNA-PK(cs)-null cells by subsequently reducing the level of Ku70. These studies demonstrate that in human cells C-NHEJ is the major DNA DSB repair pathway and they show that Ku is the critical C-NHEJ factor that regulates DNA NHEJ DSB pathway choice.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

